11119915001
Roche
RNaza, bez DNazy
from bovine pancreas
Synonim(y):
Rnase
About This Item
Polecane produkty
pochodzenie biologiczne
bovine pancreas
Poziom jakości
Formularz
solution
aktywność właściwa
≥30 units/mg protein
opakowanie
pkg of 500 μg (1 ml)
producent / nazwa handlowa
Roche
metody
DNA purification: suitable
temp. przechowywania
−20°C
Powiązane kategorie
Opis ogólny
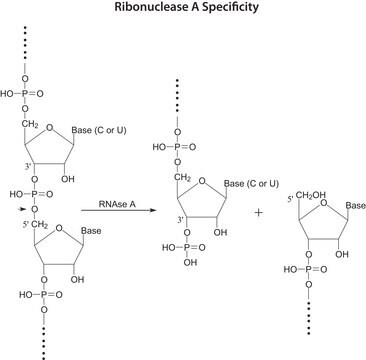
Zastosowanie
Definicja jednostki
One unit produces a decrease in absorbance at 260 nm, which is equivalent to a total conversion of RNA to oligonucleotides in one minute at +25 °C.
Postać fizyczna
Uwaga dotycząca przygotowania
- For small-scale isolation of plasmid DNA ("miniprep" from a 1.5 ml bacterial culture), use 0.5 μl of RNase, DNase-free in a reaction volume of 50 μl.
- To isolate plasmid DNA from a 100 ml bacterial culture, use 8 μl of RNase, DNase-free in a reaction volume of 2 ml.
- To isolate genomic DNA from cultured mammalian cells (5 x 107 cells), use 8 μl of RNase, DNase-free in a reaction volume of 2 ml.
Working solution: Storage and Dilution Buffer: 10 mM Tris-HCl, 5 mM CaCl2, 50% glycerol (v/v), pH 7.0.
Inne uwagi
Kod klasy składowania
12 - Non Combustible Liquids
Klasa zagrożenia wodnego (WGK)
WGK 1
Temperatura zapłonu (°F)
No data available
Temperatura zapłonu (°C)
No data available
Wybierz jedną z najnowszych wersji:
Masz już ten produkt?
Dokumenty związane z niedawno zakupionymi produktami zostały zamieszczone w Bibliotece dokumentów.
Klienci oglądali również te produkty
Protokoły
0,1 mU RNazy, wolnej od DNazy degraduje 1 μg RNA w ciągu 30 minut w temperaturze + 37 °C w objętości reakcyjnej 50 μL wody klasy PCR. Stężenie białka RNazy wolnej od DNazy wynosi 0,5 μg/μL.
0.1 mU RNase, DNase-free degrades 1 μg RNA in 30 min at + 37 °C in a reaction volume of 50 μL PCR grade water. The protein concentration of RNase, DNase-free is 0.5 μg/μL.
Nasz zespół naukowców ma doświadczenie we wszystkich obszarach badań, w tym w naukach przyrodniczych, materiałoznawstwie, syntezie chemicznej, chromatografii, analityce i wielu innych dziedzinach.
Skontaktuj się z zespołem ds. pomocy technicznej