R1003
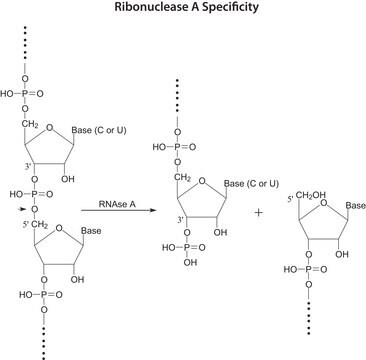
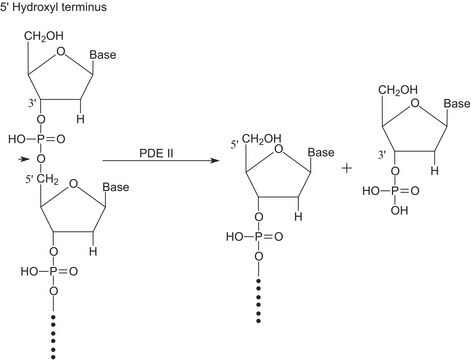
Ribonuclease T1 from Aspergillus oryzae
ammonium sulfate suspension, 300,000-600,000 units/mg protein
Sinonimo/i:
Guanyloribonuclease, Ribonucleate 3′-guanylo-oligonucleotidohydrolase
About This Item
Prodotti consigliati
Origine biologica
Aspergillus sp. (Aspergillus oryzae)
Livello qualitativo
Forma fisica
ammonium sulfate suspension
Attività specifica
300,000-600,000 units/mg protein
PM
11068 by amino acid sequence
tecniche
cell based assay: suitable
Compatibilità
suitable for separating native or denatured proteins, or nucleic acids
applicazioni
cell analysis
Temperatura di conservazione
2-8°C
Cerchi prodotti simili? Visita Guida al confronto tra prodotti
Applicazioni
Azioni biochim/fisiol
Definizione di unità
Stato fisico
Risultati analitici
Codice della classe di stoccaggio
10 - Combustible liquids
Classe di pericolosità dell'acqua (WGK)
WGK 3
Punto d’infiammabilità (°F)
Not applicable
Punto d’infiammabilità (°C)
Not applicable
Dispositivi di protezione individuale
Eyeshields, Gloves
Certificati d'analisi (COA)
Cerca il Certificati d'analisi (COA) digitando il numero di lotto/batch corrispondente. I numeri di lotto o di batch sono stampati sull'etichetta dei prodotti dopo la parola ‘Lotto’ o ‘Batch’.
Possiedi già questo prodotto?
I documenti relativi ai prodotti acquistati recentemente sono disponibili nell’Archivio dei documenti.
I clienti hanno visto anche
Articoli
Instructions for working with enzymes supplied as ammonium sulfate suspensions
Il team dei nostri ricercatori vanta grande esperienza in tutte le aree della ricerca quali Life Science, scienza dei materiali, sintesi chimica, cromatografia, discipline analitiche, ecc..
Contatta l'Assistenza Tecnica.