Wszystkie zdjęcia(2)
Key Documents
913812
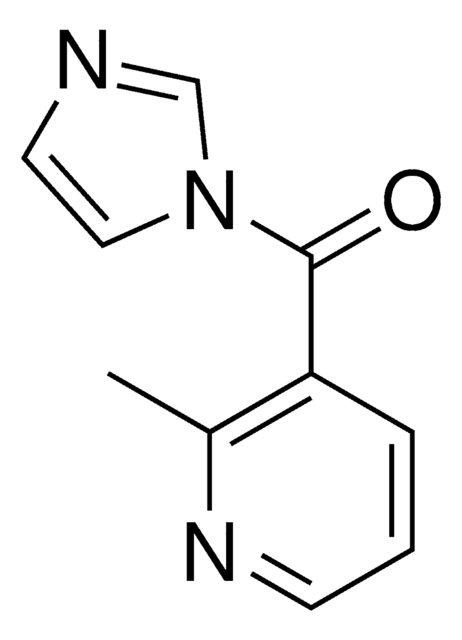
2-(Azidomethyl)nicotinic acid imidazolide
≥95%
Synonim(y):
2-(Azidomethyl)-3-(1H-imidazole-1-carbonyl)pyridine, NAI-N3, RNA SHAPE probe, icSHAPE reagent
Zaloguj sięWyświetlanie cen organizacyjnych i kontraktowych
About This Item
Wzór empiryczny (zapis Hilla):
C10H8N6O
Numer CAS:
Masa cząsteczkowa:
228.21
Numer MDL:
Kod UNSPSC:
12352106
Polecane produkty
Zastosowanie
2-(Azidomethyl)nicotinic acid imidazolide (NAI-N3) is an RNA icSHAPE probe for live-cell RNA structure profiling across the genome. icSHAPE -- or in vivo click selective 2′-hydroxyl acylation and profiling experiment -- uses NAI-N3 in a chemoaffinity method to probe RNA structure. NAI-N3 is an azido version of the cell-permeable SHAPE reagent 2-methylnicotinic acid imidazolide (NAI) that permits the tagging of NAI-N3-modified RNA with a biotin moiety for subsequent capture via streptavidin. Not only do such strategies further the understanding of RNA structure in living cells but also provide a tool for identifying regions that may be susceptible to therapeutic targeting. Recently, the azidomethylnicotinyl (AMN) group of NAI-N3 was demonstrated to block the function of gRNA and CRISPR systems, which could be reactivated by removing the AMN groups with Staudinger reduction (DPBM phosphine), overall providing a means to control nucleic acid cleavage and gene editing in live cells.
Inne uwagi
This page may contain text that has been machine translated.
Hasło ostrzegawcze
Danger
Zwroty wskazujące rodzaj zagrożenia
Zwroty wskazujące środki ostrożności
Klasyfikacja zagrożeń
Eye Irrit. 2 - Self-react. C - Skin Irrit. 2
Kod klasy składowania
5.2 - Organic peroxides and self-reacting hazardous materials
Klasa zagrożenia wodnego (WGK)
WGK 3
Temperatura zapłonu (°F)
Not applicable
Temperatura zapłonu (°C)
Not applicable
Wybierz jedną z najnowszych wersji:
Certyfikaty analizy (CoA)
Lot/Batch Number
Przepraszamy, ale COA dla tego produktu nie jest aktualnie dostępny online.
Proszę o kontakt, jeśli potrzebna jest pomoc Obsługa Klienta
Masz już ten produkt?
Dokumenty związane z niedawno zakupionymi produktami zostały zamieszczone w Bibliotece dokumentów.
Michael M Abdelsayed et al.
ACS chemical biology, 12(8), 2149-2156 (2017-07-01)
Laboratory-evolved RNAs bind a wide variety of targets and serve highly diverse functions, including as diagnostic and therapeutic aptamers. The majority of aptamers have been identified using in vitro selection (SELEX), a molecular evolution technique based on selecting target-binding RNAs
Robert C Spitale et al.
Nature, 519(7544), 486-490 (2015-03-25)
Visualizing the physical basis for molecular behaviour inside living cells is a great challenge for biology. RNAs are central to biological regulation, and the ability of RNA to adopt specific structures intimately controls every step of the gene expression program.
Fuyou Fu et al.
Scientific reports, 7(1), 2580-2580 (2017-06-02)
Macrosclereid cells, which are a layer in the seed coat of Medicago truncatula, accumulate large amounts of phytochemicals during their development. But little is known about the complex and dynamic changes during macrosclereid cell development. To characterize the phytochemicals and
Pengfei Wang et al.
PloS one, 13(11), e0207344-e0207344 (2018-11-10)
Foxtail millet is very a drought-tolerant crop. Basic helix-loop-helix (bHLH) transcription factors are involved in many drought-stress responses, but foxtail millet bHLH genes have been scarcely examined. We identified 149 foxtail millet bHLH genes in a genome-wide analysis and performed
Nasz zespół naukowców ma doświadczenie we wszystkich obszarach badań, w tym w naukach przyrodniczych, materiałoznawstwie, syntezie chemicznej, chromatografii, analityce i wielu innych dziedzinach.
Skontaktuj się z zespołem ds. pomocy technicznej
![Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine 97%](/deepweb/assets/sigmaaldrich/product/structures/179/695/86a721c8-2a4c-4e4f-bc36-6276ce7a941f/640/86a721c8-2a4c-4e4f-bc36-6276ce7a941f.png)