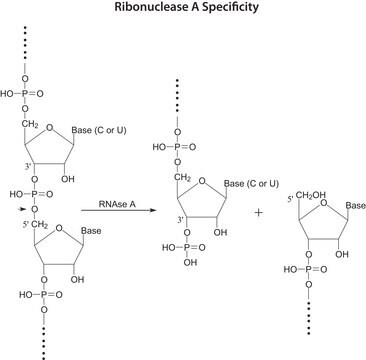
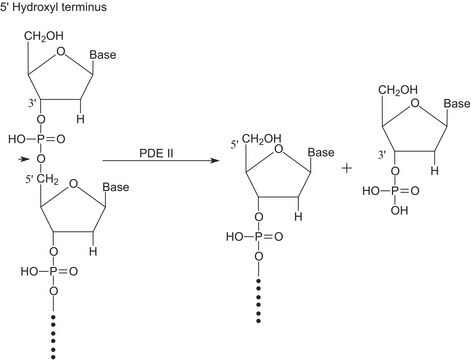
R1003
Ribonuclease T1 from Aspergillus oryzae
ammonium sulfate suspension, 300,000-600,000 units/mg protein
Sinônimo(s):
Guanyloribonuclease, Ribonucleate 3′-guanylo-oligonucleotidohydrolase
About This Item
Produtos recomendados
fonte biológica
Aspergillus sp. (Aspergillus oryzae)
Nível de qualidade
forma
ammonium sulfate suspension
atividade específica
300,000-600,000 units/mg protein
peso molecular
11068 by amino acid sequence
técnica(s)
cell based assay: suitable
adequação
suitable for separating native or denatured proteins, or nucleic acids
aplicação(ões)
cell analysis
temperatura de armazenamento
2-8°C
Procurando produtos similares? Visita Guia de comparação de produtos
Aplicação
Ações bioquímicas/fisiológicas
Definição da unidade
forma física
Nota de análise
Código de classe de armazenamento
10 - Combustible liquids
Classe de risco de água (WGK)
WGK 3
Ponto de fulgor (°F)
Not applicable
Ponto de fulgor (°C)
Not applicable
Equipamento de proteção individual
Eyeshields, Gloves
Certificados de análise (COA)
Busque Certificados de análise (COA) digitando o Número do Lote do produto. Os números de lote e remessa podem ser encontrados no rótulo de um produto após a palavra “Lot” ou “Batch”.
Já possui este produto?
Encontre a documentação dos produtos que você adquiriu recentemente na biblioteca de documentos.
Os clientes também visualizaram
Artigos
Instructions for working with enzymes supplied as ammonium sulfate suspensions
Nossa equipe de cientistas tem experiência em todas as áreas de pesquisa, incluindo Life Sciences, ciência de materiais, síntese química, cromatografia, química analítica e muitas outras.
Entre em contato com a assistência técnica