H0913
Anti-acetyl-Histone H3 (Ac-Lys9) antibody, Mouse monoclonal
clone AH3-120, purified from hybridoma cell culture
Synonym(s):
Anti-H3K9ac
About This Item
Recommended Products
biological source
mouse
Quality Level
conjugate
unconjugated
antibody form
purified immunoglobulin
antibody product type
primary antibodies
clone
AH3-120, monoclonal
form
buffered aqueous solution
mol wt
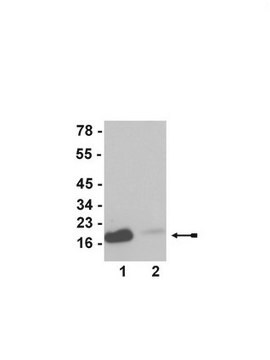
antigen ~17 kDa
species reactivity
human, bovine, Caenorhabditis elegans, frog, Drosophila, chicken, rat, mouse
packaging
antibody small pack of 25 μL
concentration
~2 mg/mL
technique(s)
immunocytochemistry: suitable
indirect ELISA: suitable
microarray: suitable
western blot: 1-2 μg/mL using whole cell extract of mouse fibroblasts 3T3 cell line treated with sodium butyrate
isotype
IgG1
UniProt accession no.
shipped in
dry ice
storage temp.
−20°C
target post-translational modification
acetylation (Lys9)
Gene Information
human ... H3F3A(3020) , H3F3B(3021) , HIST1H3A(8350) , HIST1H3B(8358) , HIST1H3C(8352) , HIST1H3D(8351) , HIST1H3E(8353) , HIST1H3F(8968) , HIST1H3G(8355) , HIST1H3H(8357) , HIST1H3I(8354) , HIST1H3J(8356) , HIST2H3A(333932) , HIST2H3C(126961) , HIST3H3(8290)
mouse ... H3f3a(15078) , H3f3b(15081) , Hist1h3a(360198) , Hist1h3b(319150) , Hist1h3c(319148) , Hist1h3d(319149) , Hist1h3e(319151) , Hist1h3f(260423) , Hist1h3g(97908) , Hist1h3h(319152) , Hist1h3i(319153) , Hist2h3b(319154) , Hist2h3c1(15077) , Hist2h3c2(97114)
rat ... H3f3b(117056)
General description
Immunogen
Application
Biochem/physiol Actions
Physical form
Disclaimer
Not finding the right product?
Try our Product Selector Tool.
related product
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 2
flash_point_f
Not applicable
flash_point_c
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Related Content
Chromatin structure plays a fundamental role in regulating processes that take place on the DNA, such as transcription, replication, and recombination, all of which occur on nucleosomal templates. Therefore, mapping the position of histones selectively modified and histone variants or non-histone components of chromatin in specific DNA sequences, can provide valuable insights into how these proteins (and their modifications) function in a chromatin context.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service