MBD0013
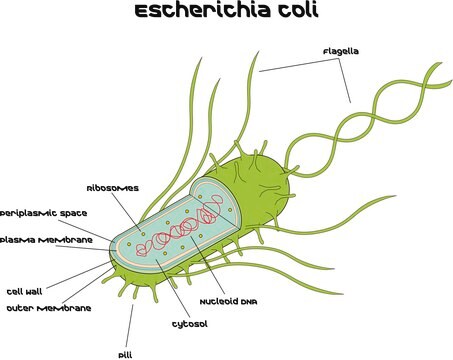
Microbial DNA standard from Escherichia coli
Suitable for PCR, sequencing and NGS, 10 ng/μL
Synonim(y):
E.Coli
About This Item
Polecane produkty
Poziom jakości
Formularz
liquid
stężenie
10 ng/μL
metody
DNA extraction: suitable
DNA sequencing: suitable
PCR: suitable
przydatność
suitable for restriction endonuclease digests, PCR amplification, Southern blots, and sequencing reactions
Warunki transportu
ambient
temp. przechowywania
−20°C
Opis ogólny
Read here how to use our standards to ensure data integrity for your microbiome research.
Zastosowanie
Suitable for Quantitative standard for PCR, Sequencing and NGS
Cechy i korzyści
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, sequencing and NGS
- Improve Bioinformatics analyses
- Increases reproducibility
- Compare results lab to lab
Postać fizyczna
Inne uwagi
Kod klasy składowania
12 - Non Combustible Liquids
Klasa zagrożenia wodnego (WGK)
WGK 1
Temperatura zapłonu (°F)
Not applicable
Temperatura zapłonu (°C)
Not applicable
Wybierz jedną z najnowszych wersji:
Certyfikaty analizy (CoA)
Nie widzisz odpowiedniej wersji?
Jeśli potrzebujesz konkretnej wersji, możesz wyszukać konkretny certyfikat według numeru partii lub serii.
Masz już ten produkt?
Dokumenty związane z niedawno zakupionymi produktami zostały zamieszczone w Bibliotece dokumentów.
Klienci oglądali również te produkty
Produkty
An overview of human microbiome research, workflow challenges, sequencing, library production, data analysis, and available microbiome reagents to support your research.
The use of standards is critical to the integrity of metagenomics research. Learn how DNA standards for bacteria, fungi, and viruses are applied to studying the microbiome. Choose standards for E. coli and other key species, as well as mixed community standards.
Nasz zespół naukowców ma doświadczenie we wszystkich obszarach badań, w tym w naukach przyrodniczych, materiałoznawstwie, syntezie chemicznej, chromatografii, analityce i wielu innych dziedzinach.
Skontaktuj się z zespołem ds. pomocy technicznej