11745824910
Roche
Biotin-Nick Translation Mix
suitable for hybridization, solution, sufficient for 40 labeling reactions, pkg of 160 μL
Synonym(s):
nick translation
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
41105500
Recommended Products
form
solution
Quality Level
usage
sufficient for 40 labeling reactions
packaging
pkg of 160 μL
manufacturer/tradename
Roche
technique(s)
hybridization: suitable
storage temp.
−20°C
General description
Convenient enzyme and nucleotide mixture.
Nick translation utilizes a combination of DNase and DNA Polymerase to nick one strand of the DNA helix, then incorporates labeled nucleotides as the polymerase examines, or "proofreads" the nicked site. The molar ratio of Biotin-16-dUTP to dTTP is adjusted to ensure that every 20th to 25th nucleotide in the newly synthesized DNA is modified with biotin. This density of haptens in the DNA yields the highest sensitivity in the immunological detection reaction.
Large plasmids, cosmids, and PCR fragments are all regularly used with this method, and the DNA can be linearized or supercoiled. Individual templates produce consistent results in the standard 90-minutes reaction, and result in an average probe length of 200 base pairs up to 500 base pairs.
Nick translation utilizes a combination of DNase and DNA Polymerase to nick one strand of the DNA helix, then incorporates labeled nucleotides as the polymerase examines, or "proofreads" the nicked site. The molar ratio of Biotin-16-dUTP to dTTP is adjusted to ensure that every 20th to 25th nucleotide in the newly synthesized DNA is modified with biotin. This density of haptens in the DNA yields the highest sensitivity in the immunological detection reaction.
Large plasmids, cosmids, and PCR fragments are all regularly used with this method, and the DNA can be linearized or supercoiled. Individual templates produce consistent results in the standard 90-minutes reaction, and result in an average probe length of 200 base pairs up to 500 base pairs.
Specificity
Heat inactivation: Stop the reaction by adding 1 μl 0.5 M EDTA (pH 8.0) and heating to 65 °C for 10 minutes.
Application
For generation of highly sensitive probes for in situ hybridization labeled with Biotin-16-dUTP.
Note: The mix can also be used for filter hybridization techniques, however, for highly sensitive filter hybridization probes, we recommend that you use Biotin-High Prime from Roche Applied Science.
For nonradioactive labeling of in situ probes with other haptens, Roche Applied Science offers the DIG-Nick Translation Mix or the Nick Translation Mix.
Note: The mix can also be used for filter hybridization techniques, however, for highly sensitive filter hybridization probes, we recommend that you use Biotin-High Prime from Roche Applied Science.
For nonradioactive labeling of in situ probes with other haptens, Roche Applied Science offers the DIG-Nick Translation Mix or the Nick Translation Mix.
Quality
Function tested in a dot blot assay.
Principle
The nick translation method is based on the ability of DNase I to introduce randomly distributed nicks into DNA at low enzyme concentrations in the presence of MgCl2. E. coli DNA Polymerase I synthesizes DNA complementary to the intact strand in a 5′ → 3′direction using the 3′-OH termini of the nick as a primer. The 5′ → 3′ exonucleolytic activity of DNA polymerase I simultaneously removes nucleotides in the direction of synthesis. The polymerase activity sequentially replaces the removed nucleotides with isotope-labeled or hapten-labeled deoxyribonucleoside triphosphates. At low temperature (+15°C), the unlabeled DNA in the reaction is thus replaced by newly synthesized labeled DNA.
Physical form
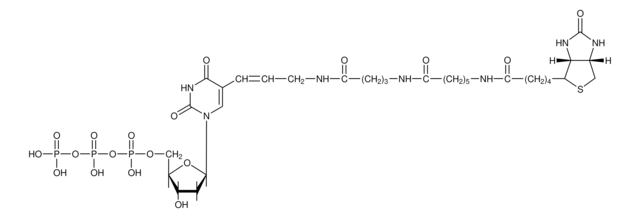
1 vial with 5x concentrated stabilized reaction buffer in 50% glycerol (v/v) and DNA Polymerase I, DNase I, 0.25mM dATP, 0.25mM dCTP, 0.25mM dGTP, 0.17mM dTTP and 0.08mM Biotin-16-dUTP.
Preparation Note
Assay Time: 100 minutes
Sample Materials
Supercoiled and linearized plasmid DNA
Supercoiled and linearized cosmid DNA
Purified PCR products
Note: Denaturing of the template before nick translation is not required.
Sample Materials
Supercoiled and linearized plasmid DNA
Supercoiled and linearized cosmid DNA
Purified PCR products
Note: Denaturing of the template before nick translation is not required.
Other Notes
For life science research only. Not for use in diagnostic procedures.
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
No data available
flash_point_c
No data available
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Shujun Zhou et al.
Breeding science, 64(1), 97-102 (2014-07-06)
Based on a recent hypothesis, "Five same genomes of endosperm are essential for its development in Lilium", it is expected that allotriploid lily (OTO) can be hybridized with diploid Oriental lily (OO) for introgression breeding in Lilium L.. To test
A Sepsi et al.
TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik, 116(6), 825-834 (2008-01-29)
In situ hybridization (multicolor GISH and FISH) was used to characterize the genomic composition of the wheat-Thinopyrum ponticum partial amphiploid BE-1. The amphiploid is a high-protein line having resistance to leaf rust (Puccinia recondita f. sp. tritici) and powdery mildew
Cornelia E Zorca et al.
Proceedings of the National Academy of Sciences of the United States of America, 112(13), E1587-E1593 (2015-03-15)
Naive CD4 T cells differentiate into several effector lineages, which generate a stronger and more rapid response to previously encountered immunological challenges. Although effector function is a key feature of adaptive immunity, the molecular basis of this process is poorly
Kristin D Kernohan et al.
MethodsX, 1, 30-35 (2014-01-01)
Emerging studies demonstrate that three-dimensional organization of chromatin in the nucleus plays a vital role in regulating the genome. DNA fluorescent in situ hybridization (FISH) is a common molecular technique used to visualize the location of DNA sequences. The vast
Shijun Li et al.
Nucleic acids research, 41(4), 2659-2672 (2013-01-12)
Titin, a sarcomeric protein expressed primarily in striated muscles, is responsible for maintaining the structure and biomechanical properties of muscle cells. Cardiac titin undergoes developmental size reduction from 3.7 megadaltons in neonates to primarily 2.97 megadaltons in the adult. This
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service