Key Documents
P5568
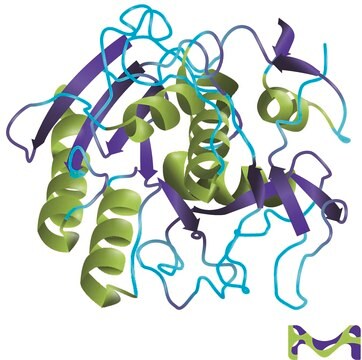
Proteinase K from Tritirachium album
≥500 units/mL, buffered aqueous glycerol solution
Synonim(y):
Endopeptidase K
About This Item
Polecane produkty
pochodzenie biologiczne
microbial (T.album
T. ALBUM)
Poziom jakości
Postać
buffered aqueous glycerol solution
masa cząsteczkowa
28.93 kDa
stężenie
≥10 mg/mL
≥500 units/mL
metody
DNA extraction: suitable
temp. przechowywania
2-8°C
Szukasz podobnych produktów? Odwiedź Przewodnik dotyczący porównywania produktów
Opis ogólny
Zastosowanie
- to break down cardiac muscle during histopathology studies
- during the digestion of HEK-293 cells
Removes endotoxins that bind to cationic proteins such as lysozyme and ribonuclease A.
Reported useful for the isolation of hepatic, yeast, and mung bean mitochondria
Determination of enzyme localization on membranes
Treatment of paraffin embedded tissue sections to expose antigen binding sites for antibody labeling.
Digestion of proteins from brain tissue samples for prions in Transmissible Spongiform Encephalopathies (TSE) research.
Działania biochem./fizjol.
Definicja jednostki
Postać fizyczna
Uwaga dotycząca przygotowania
Hasło ostrzegawcze
Danger
Zwroty wskazujące rodzaj zagrożenia
Zwroty wskazujące środki ostrożności
Klasyfikacja zagrożeń
Resp. Sens. 1
Kod klasy składowania
10 - Combustible liquids
Klasa zagrożenia wodnego (WGK)
WGK 1
Temperatura zapłonu (°F)
Not applicable
Temperatura zapłonu (°C)
Not applicable
Środki ochrony indywidualnej
Eyeshields, Faceshields, Gloves, type ABEK (EN14387) respirator filter
Certyfikaty analizy (CoA)
Poszukaj Certyfikaty analizy (CoA), wpisując numer partii/serii produktów. Numery serii i partii można znaleźć na etykiecie produktu po słowach „seria” lub „partia”.
Masz już ten produkt?
Dokumenty związane z niedawno zakupionymi produktami zostały zamieszczone w Bibliotece dokumentów.
Klienci oglądali również te produkty
Produkty
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Protokoły
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Aktywność proteinazy K mierzona za pomocą spektrofotometrii przy użyciu substratu hemoglobiny, kluczowa dla charakterystyki enzymu.
Nasz zespół naukowców ma doświadczenie we wszystkich obszarach badań, w tym w naukach przyrodniczych, materiałoznawstwie, syntezie chemicznej, chromatografii, analityce i wielu innych dziedzinach.
Skontaktuj się z zespołem ds. pomocy technicznej