推荐产品
形狀
liquid
包裝
vial of 50 μL
濃度
20 ng/μL in TE buffer; DNA (1μg of purified plasmid DNA)
技術
microbiological culture: suitable
應用
CRISPR
genome editing
啟動子
Promoter activity: constitutive
運輸包裝
dry ice
儲存溫度
−20°C
一般說明
Recent publications using CRISPR/Cas9-mediated recombineering in E. coli tout editing efficiencies near 100%, making CRISPR/Cas9-mediated recombineering the most powerful bacterial genome engineering method to date. In addition, Cas9-mediated recombineering overcomes the dependence on a second recombination step, avoids the creation of destabilizing scar sites, can be used in multiplexing, and is less time-consuming than previous protocols.
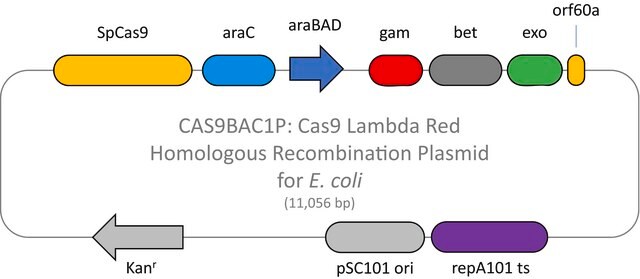
Here we present a novel dual-vector CRISPR/Cas-mediated λ-Red system for improved recombineering in E. coli. Our system is shown to facilitate homology-directed repair of DSBs created by Cas9 endonuclease, enabling genetic alterations through chromosomal integration of a donor DNA.
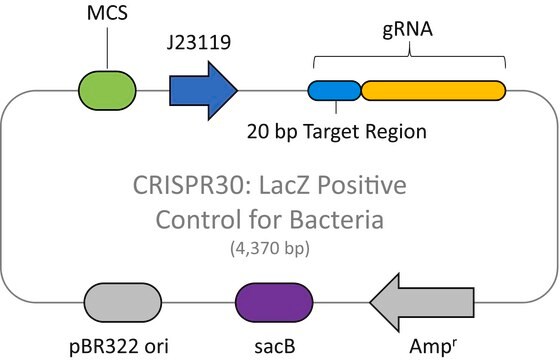
This plasmid is to be used in combination with the Cas9 Lambda Red homologous recombination plasmid for E. coli (CAS9BAC1P) as the negative control for your custom gene editing experiment. The custom gRNA (CRISPRBACD) can be designed and ordered through https://www.sigmaaldrich.com/pc/ui/genomics-home/customcrispr
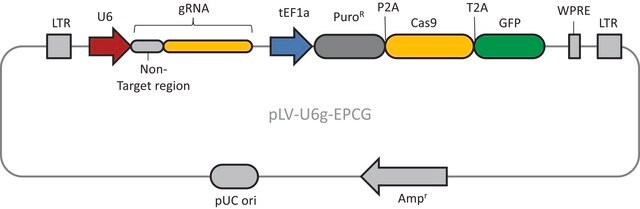
The CRISPR Non-Target Negative Control Plasmid for Bacteria (CRISPR31) contains a non-targeting spacer expressed constitutively from a J23119 promoter, a ampicillin resistance marker, a pBR322 origin of replication, and a sacB gene from Bacillus subtilis for counter-selection-based curing.
Here we present a novel dual-vector CRISPR/Cas-mediated λ-Red system for improved recombineering in E. coli. Our system is shown to facilitate homology-directed repair of DSBs created by Cas9 endonuclease, enabling genetic alterations through chromosomal integration of a donor DNA.
This plasmid is to be used in combination with the Cas9 Lambda Red homologous recombination plasmid for E. coli (CAS9BAC1P) as the negative control for your custom gene editing experiment. The custom gRNA (CRISPRBACD) can be designed and ordered through https://www.sigmaaldrich.com/pc/ui/genomics-home/customcrispr
The CRISPR Non-Target Negative Control Plasmid for Bacteria (CRISPR31) contains a non-targeting spacer expressed constitutively from a J23119 promoter, a ampicillin resistance marker, a pBR322 origin of replication, and a sacB gene from Bacillus subtilis for counter-selection-based curing.
應用
Bacterial Genome Editing
Strain Optimization
- HR-mediated recombineering for mutation or SNP analysis
- Creation of HR-mediated knock-in cell lines with promoters, fusion tags, or reporters integrated into endogenous genes
- Creation of gene knockouts in E. coli cell lines
Strain Optimization
特點和優勢
Efficient: increased efficiency of HR-mediated integration
Markerless: does not require antibiotic resistance marker insertion
Scarless: no scar sequences from marker excision which often cause off-target recombination
Multiplexing: multiple custom gRNA sequences can be used at a time
Markerless: does not require antibiotic resistance marker insertion
Scarless: no scar sequences from marker excision which often cause off-target recombination
Multiplexing: multiple custom gRNA sequences can be used at a time
原則
CRISPR/Cas systems are employed by bacteria and archaea as a defense against invading viruses and plasmids. Recently, the type II CRISPR/Cas system from the bacterium Streptococcus pyogenes has been engineered to function using two molecular components: a single Cas9 protein and a non-coding guide RNA (gRNA). The Cas9 endonuclease can be programmed with a single or dual gRNA, directing a DNA double-strand break (DSB) at a desired genomic location. Nuclease-based methods are largely toxic when employed as microbial gene editing tools because many bacteria lack the necessary DNA repair mechanisms found in eukaryotic systems. However, when CRISPR/Cas9 is used to mediate recombineering, this cytotoxic quality offers an advantage in that Cas9-induced double stranded breaks kill cells that do not recombine with the donor DNA. This provides an inherent method of selection for markerless, scarless gene editing that is dramatically more efficient and more amenable to multiplexing than traditional methods. The E. coli HR negative control plasmid (Catalog Number CRISPR31-1UG) contains a gRNA sequence targeting no known genomic DNA seqeuence in Wild-type E. Coli. This makes it suitable for use a negative control when used in conjunction with CAS9BAC1P-1UG.
儲存類別代碼
12 - Non Combustible Liquids
水污染物質分類(WGK)
WGK 2
閃點(°F)
Not applicable
閃點(°C)
Not applicable
Yifan Li et al.
Metabolic engineering, 31, 13-21 (2015-07-05)
Engineering cellular metabolism for improved production of valuable chemicals requires extensive modulation of bacterial genome to explore complex genetic spaces. Here, we report the development of a CRISPR-Cas9 based method for iterative genome editing and metabolic engineering of Escherichia coli.
Michael E Pyne et al.
Applied and environmental microbiology, 81(15), 5103-5114 (2015-05-24)
To date, most genetic engineering approaches coupling the type II Streptococcus pyogenes clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system to lambda Red recombineering have involved minor single nucleotide mutations. Here we show that procedures for carrying out more complex
商品
In this article, we present an application of our novel E. coli CRISPR/Cas-mediated Lambda-Red (λ-Red) homologous recombination (HR) vector system, which facilitates gene editing through the homology-directed repair (HDR) of double-stranded DNA breaks (DSBs) created by Cas9 endonuclease, using either ssDNA or dsDNA as an editing template.
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门