Wichtige Dokumente
R1003
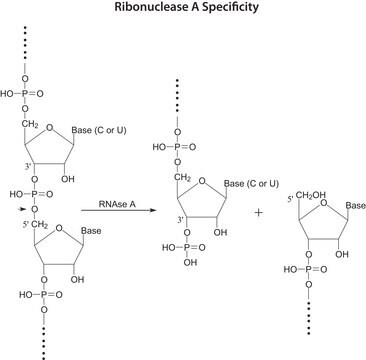
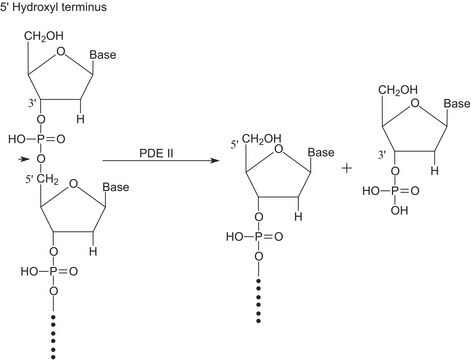
Ribonuklease T1 aus Aspergillus oryzae
ammonium sulfate suspension, 300,000-600,000 units/mg protein
Synonym(e):
Guanyloribonuclease, Ribonucleate 3′-guanylo-oligonucleotidohydrolase
About This Item
Empfohlene Produkte
Biologische Quelle
Aspergillus sp. (Aspergillus oryzae)
Qualitätsniveau
Form
ammonium sulfate suspension
Spezifische Aktivität
300,000-600,000 units/mg protein
Mol-Gew.
11068 by amino acid sequence
Methode(n)
cell based assay: suitable
Eignung
suitable for separating native or denatured proteins, or nucleic acids
Anwendung(en)
cell analysis
Lagertemp.
2-8°C
Suchen Sie nach ähnlichen Produkten? Aufrufen Leitfaden zum Produktvergleich
Anwendung
Biochem./physiol. Wirkung
Einheitendefinition
Physikalische Form
Hinweis zur Analyse
Lagerklassenschlüssel
10 - Combustible liquids
WGK
WGK 3
Flammpunkt (°F)
Not applicable
Flammpunkt (°C)
Not applicable
Persönliche Schutzausrüstung
Eyeshields, Gloves
Hier finden Sie alle aktuellen Versionen:
Besitzen Sie dieses Produkt bereits?
In der Dokumentenbibliothek finden Sie die Dokumentation zu den Produkten, die Sie kürzlich erworben haben.
Kunden haben sich ebenfalls angesehen
Artikel
Instructions for working with enzymes supplied as ammonium sulfate suspensions
Unser Team von Wissenschaftlern verfügt über Erfahrung in allen Forschungsbereichen einschließlich Life Science, Materialwissenschaften, chemischer Synthese, Chromatographie, Analytik und vielen mehr..
Setzen Sie sich mit dem technischen Dienst in Verbindung.