SRP2043
PPAR, α human
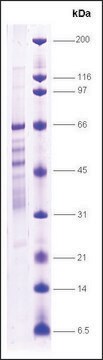
recombinant, expressed in E. coli, ≥75% (SDS-PAGE)
Sinonimo/i:
MGC2237, MGC2452, NR1C1, PPAR, PPARalpha, hPPAR
About This Item
Prodotti consigliati
Origine biologica
human
Ricombinante
expressed in E. coli
Saggio
≥75% (SDS-PAGE)
Forma fisica
frozen liquid
PM
~54 kDa
Confezionamento
pkg of 10 μg
Condizioni di stoccaggio
avoid repeated freeze/thaw cycles
Concentrazione
250 μg/mL
tecniche
electrophoretic mobility shift assay: suitable
Colore
clear colorless
N° accesso NCBI
N° accesso UniProt
Condizioni di spedizione
dry ice
Temperatura di conservazione
−70°C
Informazioni sul gene
human ... PPARA(5465)
Azioni biochim/fisiol
Stato fisico
Nota sulla preparazione
Codice della classe di stoccaggio
10 - Combustible liquids
Classe di pericolosità dell'acqua (WGK)
WGK 1
Punto d’infiammabilità (°F)
Not applicable
Punto d’infiammabilità (°C)
Not applicable
Certificati d'analisi (COA)
Cerca il Certificati d'analisi (COA) digitando il numero di lotto/batch corrispondente. I numeri di lotto o di batch sono stampati sull'etichetta dei prodotti dopo la parola ‘Lotto’ o ‘Batch’.
Possiedi già questo prodotto?
I documenti relativi ai prodotti acquistati recentemente sono disponibili nell’Archivio dei documenti.
Il team dei nostri ricercatori vanta grande esperienza in tutte le aree della ricerca quali Life Science, scienza dei materiali, sintesi chimica, cromatografia, discipline analitiche, ecc..
Contatta l'Assistenza Tecnica.