94403
Hoechst 33258 solution
1 mg/mL in H2O, ≥98.0% (HPLC)
Sinônimo(s):
bisBenzimide H 33258 solution
About This Item
Produtos recomendados
Ensaio
≥98.0% (HPLC)
Formulário
liquid
concentração
1 mg/mL in H2O
solubilidade
DMF: soluble
H2O: soluble
fluorescência
λex 355 nm; λem 465 nm in TE buffer; DNA
temperatura de armazenamento
2-8°C
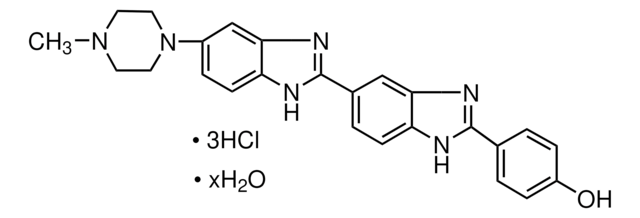
cadeia de caracteres SMILES
Cl.Cl.Cl.CN1CCN(CC1)c2ccc3[nH]c(nc3c2)-c4ccc5nc([nH]c5c4)-c6ccc(O)cc6
InChI
1S/C25H24N6O.3ClH/c1-30-10-12-31(13-11-30)18-5-9-21-23(15-18)29-25(27-21)17-4-8-20-22(14-17)28-24(26-20)16-2-6-19(32)7-3-16;;;/h2-9,14-15,32H,10-13H2,1H3,(H,26,28)(H,27,29);3*1H
chave InChI
SMNPLAKEGAEPJD-UHFFFAOYSA-N
Descrição geral
Aplicação
Código de classe de armazenamento
10 - Combustible liquids
Ponto de fulgor (°F)
Not applicable
Ponto de fulgor (°C)
Not applicable
Escolha uma das versões mais recentes:
Já possui este produto?
Encontre a documentação dos produtos que você adquiriu recentemente na biblioteca de documentos.
Os clientes também visualizaram
Nossa equipe de cientistas tem experiência em todas as áreas de pesquisa, incluindo Life Sciences, ciência de materiais, síntese química, cromatografia, química analítica e muitas outras.
Entre em contato com a assistência técnica