S7822-M
CpGenome Universal Unmethylated DNA
Kit containing two separate unmethylated genomic DNA controls for gene methylation studies and methylation-specific PCR (MSP).
Sinônimo(s):
Unmethylated DNA control
Faça loginpara ver os preços organizacionais e de contrato
About This Item
Código UNSPSC:
12161503
eCl@ss:
32161000
NACRES:
NA.25
Produtos recomendados
Nível de qualidade
reatividade de espécies
human
fabricante/nome comercial
Chemicon®
CpGenome
aplicação(ões)
genomic analysis
Categorias relacionadas
Descrição geral
CpGenome Universal Unmethylated DNA Set provides 2 separate genomic DNA controls for gene methylation studies. This product is intended for use with the CpGenome DNA Modification Kit (S7820) and can be subsequently used as an unmethylated DNA control for gene methylation studies.
Materials Provided:
Universal Unmethylated DNA Vial A contains 5 micrograms (50μL) of human genomic DNA at a concentration of 0.1 μg/μL. Universal Unmethylated DNA Vial B contains 5 micrograms (50μL), of genomic DNA isolated from a primary human fetal cell line at a concentration of 0.1 μg/μL. Note: The DNA must first be bisulfite modified using (S7820) to use a specific primer set for the unmethylated form of the gene of interest.
Validation:
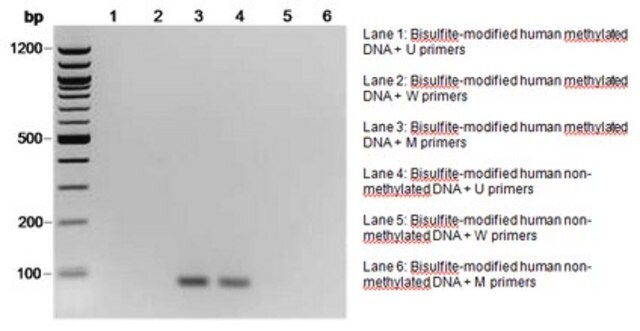
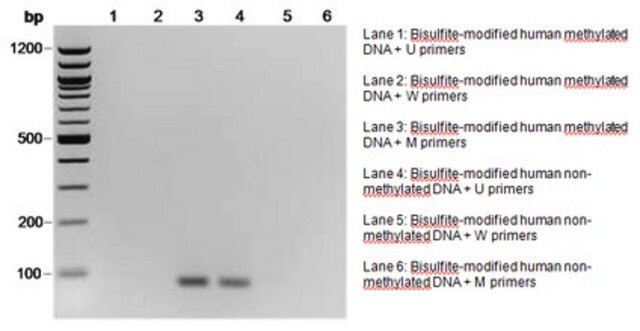
Methylation-specific PCR (MSP) was performed on the DNA following bisulfite modification. Various primer sets from the CpG WIZ product line were used in the assay. DNA from Vial A has been validated with primers from several CpG WIZ amplification kits. Vial B DNA is also unmethylated at several promoters assayed with the CpG WIZ kits and is supplied as an alternative source of DNA for promoters that may be partially methylated in Vial A DNA.
CpG Genome and CpG WIZ Methylation Products apply technologies exclusively licensed from The Johns Hopkins University School of Medicine. Methylation-specific PCR (MSP) technology is covered by U.S. Patent # 5,786,146.
Materials Provided:
Universal Unmethylated DNA Vial A contains 5 micrograms (50μL) of human genomic DNA at a concentration of 0.1 μg/μL. Universal Unmethylated DNA Vial B contains 5 micrograms (50μL), of genomic DNA isolated from a primary human fetal cell line at a concentration of 0.1 μg/μL. Note: The DNA must first be bisulfite modified using (S7820) to use a specific primer set for the unmethylated form of the gene of interest.
Validation:
Methylation-specific PCR (MSP) was performed on the DNA following bisulfite modification. Various primer sets from the CpG WIZ product line were used in the assay. DNA from Vial A has been validated with primers from several CpG WIZ amplification kits. Vial B DNA is also unmethylated at several promoters assayed with the CpG WIZ kits and is supplied as an alternative source of DNA for promoters that may be partially methylated in Vial A DNA.
CpG Genome and CpG WIZ Methylation Products apply technologies exclusively licensed from The Johns Hopkins University School of Medicine. Methylation-specific PCR (MSP) technology is covered by U.S. Patent # 5,786,146.
Methylation of cytosines located 5′ to guanosine is known to have a profound effect on the expression of many eukaryotic genes. In normal cells methylation occurs predominantly in CG-poor regions, while CG-rich areas, called CpG-islands remain unmethylated. The exceptions are the extensive methylation of CpG islands associated with transcriptional inactivation of regulatory regions of imprinted genes and genes on the inactive X-chromosome of females. Aberrant methylation of normally unmethylated CpG islands has been documented as a relatively frequent event in immortalized and transformed cells and has been associated with transcriptional inactivation of defined tumor suppresser genes in human cancers. Hundreds of CpG islands are now known to exhibit the characteristic of hypermethylation in tumors.
Several methods have been developed to determine the methylation status of cytosine. These include digestion with methylation sensitive restriction enzymes as in restriction landmark genomic scanning, oligonucleotide arrays, genomic DNA sequencing and methylation specific PCR (MSP). Some techniques are more useful for discovery while others are better used for monitoring of known methylated cytosines. Genomic DNA sequencing, although time consuming and labor intensive, offers a more universal detection method. MSP is now an established technology for the monitoring of abnormal gene methylation in selected gene sequences. Utilizing small amounts of DNA, this procedure offers sensitive and specific detection of 5-methylcytosine in promoters. It is being exploited to define tumor suppresser gene function, and to provide a new strategy for early tumor detection.
The initial step of both genomic sequencing and MSP is to perform a bisulfite modification of the DNA sample. MSP then involves PCR amplification with specific primers designed to distinguish methylated from unmethylated DNA.
Several methods have been developed to determine the methylation status of cytosine. These include digestion with methylation sensitive restriction enzymes as in restriction landmark genomic scanning, oligonucleotide arrays, genomic DNA sequencing and methylation specific PCR (MSP). Some techniques are more useful for discovery while others are better used for monitoring of known methylated cytosines. Genomic DNA sequencing, although time consuming and labor intensive, offers a more universal detection method. MSP is now an established technology for the monitoring of abnormal gene methylation in selected gene sequences. Utilizing small amounts of DNA, this procedure offers sensitive and specific detection of 5-methylcytosine in promoters. It is being exploited to define tumor suppresser gene function, and to provide a new strategy for early tumor detection.
The initial step of both genomic sequencing and MSP is to perform a bisulfite modification of the DNA sample. MSP then involves PCR amplification with specific primers designed to distinguish methylated from unmethylated DNA.
Embalagem
2 x 5 μg
forma física
Liquid in TE (10mM Tris-HCL, 0.1mM EDTA) with no preservatives.
Armazenamento e estabilidade
Recommended Storage: -15°C to -25°C, in aliquots, protected from freeze-thaws for up to two years from date of receipt. Do not freeze and thaw the same -20°C aliquot more than 3X for best results. Multiple freeze thaws can fragment DNA. Storage at -70°C can be used for longer term storage if necessary; minimize multiple freeze thaws for best results.
Informações legais
CHEMICON is a registered trademark of Merck KGaA, Darmstadt, Germany
Exoneração de responsabilidade
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Código de classe de armazenamento
12 - Non Combustible Liquids
Classe de risco de água (WGK)
WGK 1
Ponto de fulgor (°F)
Not applicable
Ponto de fulgor (°C)
Not applicable
Certificados de análise (COA)
Busque Certificados de análise (COA) digitando o Número do Lote do produto. Os números de lote e remessa podem ser encontrados no rótulo de um produto após a palavra “Lot” ou “Batch”.
Já possui este produto?
Encontre a documentação dos produtos que você adquiriu recentemente na biblioteca de documentos.
Os clientes também visualizaram
The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women.
Jyothi S Prabhu,Kanu Wahi,Aruna Korlimarla,Marjorrie Correa,Suraj Manjunath,N Raman et al.
Tumour Biology : the Journal of the International Society For Oncodevelopmental Biology and Medicine null
Nossa equipe de cientistas tem experiência em todas as áreas de pesquisa, incluindo Life Sciences, ciência de materiais, síntese química, cromatografia, química analítica e muitas outras.
Entre em contato com a assistência técnica