MBD0013
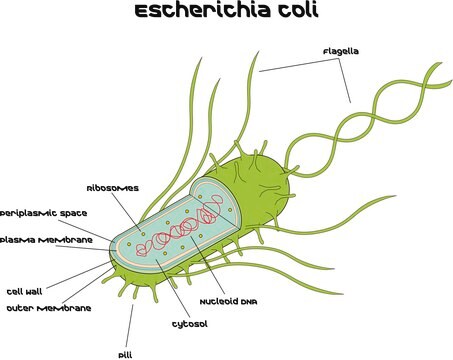
Microbial DNA standard from Escherichia coli
Suitable for PCR, sequencing and NGS, 10 ng/μL
Synonym(e):
E.Coli
About This Item
Empfohlene Produkte
Qualitätsniveau
Form
liquid
Konzentration
10 ng/μL
Methode(n)
DNA extraction: suitable
DNA sequencing: suitable
PCR: suitable
Eignung
suitable for restriction endonuclease digests, PCR amplification, Southern blots, and sequencing reactions
Versandbedingung
ambient
Lagertemp.
−20°C
Allgemeine Beschreibung
Read here how to use our standards to ensure data integrity for your microbiome research.
Anwendung
Suitable for Quantitative standard for PCR, Sequencing and NGS
Leistungsmerkmale und Vorteile
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, sequencing and NGS
- Improve Bioinformatics analyses
- Increases reproducibility
- Compare results lab to lab
Physikalische Form
Sonstige Hinweise
Lagerklassenschlüssel
12 - Non Combustible Liquids
WGK
WGK 1
Flammpunkt (°F)
Not applicable
Flammpunkt (°C)
Not applicable
Analysenzertifikate (COA)
Suchen Sie nach Analysenzertifikate (COA), indem Sie die Lot-/Chargennummer des Produkts eingeben. Lot- und Chargennummern sind auf dem Produktetikett hinter den Wörtern ‘Lot’ oder ‘Batch’ (Lot oder Charge) zu finden.
Besitzen Sie dieses Produkt bereits?
In der Dokumentenbibliothek finden Sie die Dokumentation zu den Produkten, die Sie kürzlich erworben haben.
Kunden haben sich ebenfalls angesehen
Artikel
An overview of human microbiome research, workflow challenges, sequencing, library production, data analysis, and available microbiome reagents to support your research.
The use of standards is critical to the integrity of metagenomics research. Learn how DNA standards for bacteria, fungi, and viruses are applied to studying the microbiome. Choose standards for E. coli and other key species, as well as mixed community standards.
Unser Team von Wissenschaftlern verfügt über Erfahrung in allen Forschungsbereichen einschließlich Life Science, Materialwissenschaften, chemischer Synthese, Chromatographie, Analytik und vielen mehr..
Setzen Sie sich mit dem technischen Dienst in Verbindung.