Cell Viability and Proliferation Assays
- DNA Synthesis Proliferation Assays
- Metabolic Proliferation Assays
- Luminescent Cell Viability Assays
- Fluorescent Dye Proliferation Assays
- 3D Cell Culture- Live/Dead/Total Cell Triple Staining
- Trypan Blue Cell Counting
Introduction
Assays to measure cellular proliferation, cell viability, and cytotoxicity are commonly used to monitor the response and health of cells in culture after treatment with various stimuli. The proper choice of an assay method depends on the number and type of cells used as well as the expected outcome. Assays for cell proliferation may monitor the number of cells over time, the number of cellular divisions, metabolic activity, or DNA synthesis. Cell counting using viability dyes such as trypan blue or Calcein-AM can provide both the rate of proliferation as well as the percentage of viable cells.
Overview of Cell Viability and Proliferation Assays
DNA Synthesis Proliferation Assays
BrdU Cell Proliferation Assay
Cell proliferation may be studied by monitoring the incorporation of a radioisotope, [3H]-thymidine, into cellular DNA, followed by autoradiography. Alternatively, 5-bromo-2′-deoxy-uridine (BrdU assays) may be used instead of thymidine. Cells that have incorporated BrdU into their DNA are easily detected using a monoclonal antibody against BrdU and an enzyme- or fluorochrome-conjugated secondary antibody.

Figure 1.A, Proliferating cells in the eye of E4 chick embryo using BrdU staining B, Anti-BrdU antibody validation using camptothecin. By treating Jurkat cells with cell cycle arrest agent camptothecin, circulating cells are trapped at the G1/S phase transition.
EdU Proliferation Assays
Baseclick EdU proliferation assays provide an efficient method for fluorescence detection of replicating DNA. The modified nucleoside EdU is added to live cells and is incorporated into replicating DNA. Cu-induced click chemistry allows rapid attachment of fluorescent probes to the EdU. This provides for a quantitative way to monitor cells that are proliferating. The assays are available in various formats for microscopic imaging, flow cytometry, high throughput screening, and for in vivo experiments. Four different fluorescent probes with peak excitation of 488, 555, 594, and 647 are available for multiplexing capability with other fluorescent probes.

Figure 2.Edu-Click cell proliferation kits incorporate EdU (5-ethynyl-2’-deoxyuridine) into DNA during active DNA synthesis and are measured using a click-chemistry fluorescent detection method.
Metabolic Proliferation Assays
Assays that measure metabolic activity are suitable for analyzing proliferation, viability, and cytotoxicity. The reduction of tetrazolium salts such as MTT, XTT, and WST-1 to colored formazan compounds or the bioreduction of resazurin occurs only in metabolically active cells. Actively proliferating cells increase their metabolic activity, while cells exposed to toxins will have decreased activity.
MTT Cell Proliferation Assays
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5- diphenyltetrazolium bromide; thiazolyl blue) is a water soluble tetrazolium salt yielding a yellowish solution when prepared in media or salt solutions lacking phenol red. Dissolved MTT is converted to an insoluble purple formazan by cleavage of the tetrazolium ring by dehydrogenase enzymes. This water-insoluble formazan can be solubilized using isopropanol or other solvents, and the dissolved material is measured spectrophotometrically using absorbance as a function of concentration of converted dye.
XTT Cell Proliferation Assays
In contrast to MTT, the cleavage product of XTT is soluble in water; a solubilization step is therefore not required. The tetrazolium salt XTT is cleaved to formazan by a complex cellular mechanism. This bioreduction occurs in viable cells only, and is related to NAD(P)H production through glycolysis. The amount of formazan dye formed directly correlates to the number of metabolically active cells in the culture.
WST-1 Cell Proliferation Assays
The stable tetrazolium salt WST-1 is cleaved to a soluble formazan by a complex cellular mechanism that occurs primarily at the cell surface. This bioreduction is largely dependent on the glycolytic production of NAD(P)H in viable cells. The amount of formazan dye formed directly correlates to the number of metabolically active cells in the culture.
Protocol Guide: WST-1 Assay for Cell Viability and Proliferation. Protocols, FAQs and troubleshooting

Figure 3.The MTT assay is a colorimetric assay for assessing cell proliferation based on metabolic activity. NAD(P)H-dependent cellular oxidoreductase enzymes reflect the number of viable cells present. These enzymes are capable of reducing the yellow tetrazolium dye MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide to its insoluble formazan, which has a purple color.
Luminescent Cell Viability Assays
Because ATP is an indicator of metabolically active cells, the number of viable cells can be assessed based on the amount of ATP available. The ATP Cell Viability Luciferase Assay offers a highly sensitive homogenous assay for quantifying ATP in cell cultures. This kit employs firefly luciferase to oxidize D-Luciferin and the resulting production of light to assess the amount of ATP available in cell cultures. The sensitive assay procedure involves a single addition of ATP detection cocktail directly to cells cultured in a serum-supplemented medium. No cell washing, medium removal, or multiple pipetting are required. The kit is sensitive enough for detection of a single cell or 0.01 picomoles of ATP. The signal produced is linear within six orders of magnitude. By relating the amount of ATP to the number of viable cells, the assay has wide applications, ranging from the determination of viable cell numbers to cell proliferation to cell cytotoxicity.

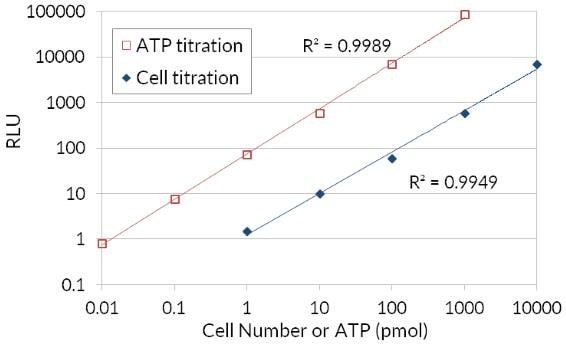
Figure 4.Bioluminescent ATP luciferase cell viability assay. Firefly luciferase’s use of ATP to oxidize D-Luciferin and the resulting production of light in order to assess the amount of ATP available that correlates to cell number and viability.
Fluorescent Dye Proliferation Assays
CFSE Labeling
5(6)-Carboxyfluorescein diacetate N-succinimidyl ester (CFSE) is a popular choice for measuring the number of divisions undergone by a cellular population. Upon entering the cell, CFSE is cleaved by intracellular esterases to form the fluorescent compound and the succinimidyl ester group covalently reacts with primary amines on intracellular proteins. Upon division, the fluorescence intensity of each daughter cell is halved enabling simple detection of the number of cell divisions by flow cytometry. CFSE has been widely used to measure the proliferation of lymphocytes, including T cells.
Live/Dead Cell Double Staining
Live/Dead Cell Double Staining can be utilized for simultaneous fluorescence detection of viable and dead cells. /US/enCalcein-AM is a highly lipophilic and cell membrane-permeable dye. Though /US/enCalcein-AM itself is not a fluorescent molecule, the calcein generated from /US/enCalcein-AM by esterase in a viable cell emits a strong green fluorescence (λex 490 nm, λem 515 nm). /US/enCalcein-AM therefore only stains viable cells. In contrast, the nuclei-staining dye Propidium Iodine cannot pass through a viable cell membrane. It reaches the nucleus by passing through disordered areas of dead cell membrane, and intercalates with the DNA double helix to emit red fluorescence (λex 535 nm, λem 617 nm). Since both Calcein and PI-DNA can be excited with 490 nm light, simultaneous monitoring of viable and dead cells is possible with a single-excitation fluorescence microscope.

Figure 5.Live/Dead Cell Double Staining
3D Cell Culture- Live/Dead/Total Cell Triple Staining
The Cell Viability Imaging Kit is a three-color assay that can be used with 2D and 3D cell cultures for simultaneous fluorescence staining of viable cells (Calcein-AM), dead cells (Propidium Iodide/PI), as well as total cells (Hoechst 33342).
- Calcein-AM fluoresces green on binding calcium, relying on esterase activity present only in metabolically-active viable cells.
- Propidium Iodide (PI) is a nuclear dye that is excluded by the membrane of live cells, but passes through the damaged membrane of dead cells, intercalating with the DNA to emit a strong red fluorescence.
- Hoechst 33342 is a DNA staining dye that exhibits low cytotoxicity. It fluoresces blue and is used as an indicator of total cell count.
Trypan Blue Cell Counting
Trypan Blue is one of several stains recommended for use in dye exclusion procedures for viable cell counting. This method is based on the principle that live (viable) cells do not take up the blue dye, whereas dead (non-viable) cells do. Cell viability can be calculated using the ratio of total live/total cells (live and dead). Staining also facilitates the visualization of overall cell morphology.
NOTE: Trypan Blue has a greater affinity for serum proteins than for cellular protein. If the background is too dark due to the presence of serum in the matrix, cells should be pelleted and resuspended in protein-free medium or salt solution prior to counting.

Figure 6.Cell Counting Using a Hemocytometer and Trypan Blue
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?