N5661
Nuclease S1 from Aspergillus oryzae
for single-strand DNA/RNA digestion
Sinónimos:
Endonuclease S1
Iniciar sesiónpara Ver la Fijación de precios por contrato y de la organización
About This Item
Número de CAS:
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Productos recomendados
biological source
Aspergillus sp. (A. oryzae)
form
solution
concentration
≥100000 units/mL
technique(s)
DNA purification: suitable
suitability
suitable for nucleic acid purification
application(s)
cell analysis
shipped in
wet ice
storage temp.
−20°C
¿Está buscando productos similares? Visita Guía de comparación de productos
Categorías relacionadas
General description
The Nuclease S1 enzyme from Aspergillus oryzae has the ability to degrade single-stranded oligonucleotides composed of either deoxynucleotides or ribonucleotides.
Application
Nuclease S1 from Aspergillus oryzae has been used in a study to assess a biochemical method for mapping mutational alterations in DNA. It has also been used in a study to investigate the DNA damage and repair in a γ-irradiated rat brain tumor.
Biochem/physiol Actions
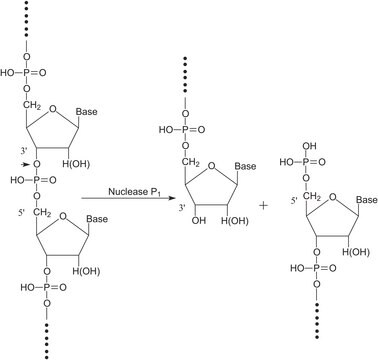
Nuclease S1 isolated from Aspergillus oryzae exhibits endo- and exolytic hydrolytic activity for the phosphodiester bonds of single-stranded DNA and RNA yielding 5′-phosphomononucleotide and 5′-phosphooligonucleotide end-products. It is used to digest non-annealed polynucleotide tails and hairpin loops in RNA and DNA duplexes and can be used to convert superhelical DNA to the linear form.
SI nuclease from Aspergillus oryzae can generate double-stranded DNA breaks in response to DNA nicks or abasic sites.
Unit Definition
One unit will cause 1.0 microgram of single-stranded nucleic acid to become perchloric acid soluble per minute at pH 4.6 at 37°C.
Physical form
Solution containing 30 mM sodium acetate, 50 mM NaCl, 1 mM ZnCl2, 50% glycerol, 2 mg/ml protein
Solo componentes del kit
Referencia del producto
Descripción
- 30mM Sodium acetate .25-.25 %
- 50mM Sodium chloride .29 %
- 1mM Zinc chloride .01 %
- Glycerol 50 %
- 2mg/mL Protein .2 %
Storage Class
10 - Combustible liquids
wgk_germany
WGK 2
flash_point_f
Not applicable
flash_point_c
Not applicable
Elija entre una de las versiones más recientes:
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
F Harada et al.
Nucleic acids research, 2(6), 865-871 (1975-06-01)
Nuclease S1 specifically hydrolizes tRNAs in their anticodon loops, forming new 5' phosphate and 3' OH ends. Some single-stranded regions are not cut by nuclease S1. The strong preference of nuclease S1 for the anticodon region can be used for
M A Chaudhry et al.
Nucleic acids research, 23(19), 3805-3809 (1995-10-11)
Defined DNA substrates containing discrete abasic sites or paired abasic sites set 1, 3, 5 and 7 bases apart on opposite strands were constructed to examine the reactivity of S1, mung bean and P1 nucleases towards abasic sites. None of
P Beard et al.
Journal of virology, 12(6), 1303-1313 (1973-12-01)
S(1) nuclease, the single-strand specific nuclease from Aspergillus oryzae can cleave both strands of circular covalently closed, superhelical simian virus 40 (SV40) DNA to generate unit length linear duplex molecules with intact single strands. But circular, covalently closed, nonsuperhelical DNA
T E Shenk et al.
Proceedings of the National Academy of Sciences of the United States of America, 72(3), 989-993 (1975-03-01)
S1 nuclease (EC 3.1.4.X), a single-strand-specific nuclease, can be used to accurately map the location of mutational alterations in simian virus 40 (SV40) DNA. Deletions of between 32 and 190 base pairs, which are at or below the limit of
S1 nuclease from Aspergillus oryzae for the detection of DNA damage and repair in the gamma-irradiated intracerebral rat gliosarcoma 9L.
P H Gutin et al.
Radiation research, 72(1), 100-106 (1977-10-01)
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico