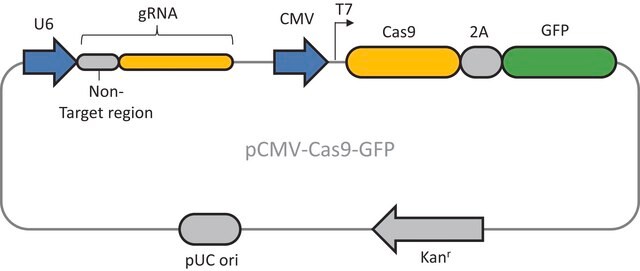
CRISPR02
CRISPR Nickase EMX1 Positive Control
About This Item
Productos recomendados
form
liquid
Quality Level
packaging
pkg of 3 vials (50μL aliquot for each of the 3 kit components)
concentration
20 ng/μL in TE buffer; DNA (1μg of plasmid DNA)
application(s)
CRISPR
shipped in
dry ice
storage temp.
−20°C
Categorías relacionadas
General description
Application
- Creation of gene knockouts in cell lines
- Creation of knock-in cell lines with promoters, fusion tags or reporters integrated into endogenous genes
Features and Benefits
Components
Principle
Physical form
Preparation Note
Other Notes
Legal Information
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Artículos
CRISPR endonucleases have shown wide variation in their activity, even among multiple CRISPRs designed within close genomic proximity.
View experimental data showing crispr/cas expression and enrichment using FACS
Protocolos
Learn about CRISPR Cas9, what it is and how it works. CRISPR is a new, affordable genome editing tool enabling access to genome editing for all.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico