추천 제품
생물학적 소스
bacterial (marine bacterium BMTU 3346)
Quality Level
재조합
expressed in E. coli
형태
solution
포장
pkg of 100 U (11775367001)
pkg of 500 U (11775375001)
제조업체/상표
Roche
농도
1000 U/mL
기술
activity assay: suitable
색상
colorless
최적 pH
8.3-8.9
solubility
water: soluble
적합성
suitable for enzyme test
응용 분야
life science and biopharma
외래 활성
DNA decontamination:, present ( >= 10 e5 copies )
RNases with MS- II- RNA ≤10 units, none detected
Unspec.nucleases w. lambda-DNA 20 units, none detected
Unspec.nucleases w.M13mp9ss-DNA ≤20 units, none detected
Unspec.nucleases w.pBR 322-DNA ≤20 units, none detected
저장 온도
−20°C
관련 카테고리
일반 설명
Enzyme Characteristics
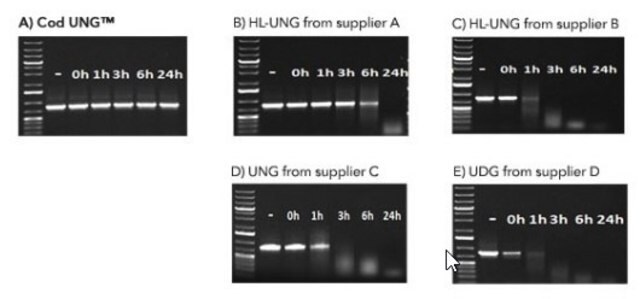
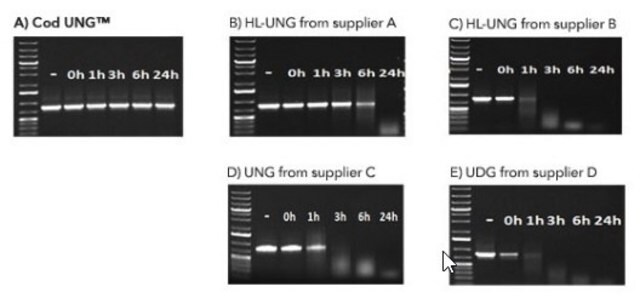
The BMTU 3346 enzyme is inactivated more quickly (2 minutes at +95°C) than the corresponding enzyme from E. coli (10 minutes at +95°C). It has also been reported that UNG from E. coli remains partially active, leading to the degradation of the dU-containing PCR product. In contrast, the heat-labile UNG does not degrade dU-PCR products within at least several hours of incubation at +2 to +8°C. Therefore, it is not necessary to freeze the PCR product immediately after amplification or to hold the reaction mixture at -70°C.
특이성
- Uracil-DNA glycosylase hydrolyzes uracil-glycosidic bonds at U-DNA sites in single- and doublestranded DNA, excising uracil and creating alkali sensitive abasic sites in the DNA.
- The enzyme is active on both single-stranded DNA and double-standed DNA.
- Uracil-DNA glycosylase is inactive on RNA and native, uracil-free DNA.
- Since uracil-DNA glycosylase has no metal ion requirements, it is fully active in the presence of EDTA.
Heat inactivation: 95 °C for 2 min
애플리케이션
Important Note: For highly sensitive techniques like real-time PCR we recommend our LightCycler® Uracil-DNA Glycosylase which is optimized for this application.
특징 및 장점
- Prevent carryover contamination in PCR.
- Increase the efficiency of site-directed mutagenesis procedures.
- Label oligonucleotide probes.
- Perform faster inactivation than the corresponding enzyme from E. coli due to the lower thermostability of the enzyme.
- Obtain no degradation of dU-PCR products within at least several hours of incubation at +2 to +8°C.
Contents
The enzyme is supplied as 1 U/μl solution in storage buffer.
품질
The enzyme does not contain any contaminating exo- or endonucleases and is free from RNase activity, according to the current quality control procedures.
단위 정의
One Lindahl unit is defined as the amount of enzyme necessary to release 1 mol uracil at +37 °C in 1 minute. One Lindahl unit is comparable to 520 000 units based on our unit definition.
Volume Activity: 1 U/μl
기타 정보
법적 정보
유해 및 위험 성명서
예방조치 성명서
Hazard Classifications
Aquatic Chronic 3
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 2
Flash Point (°F)
does not flash
Flash Point (°C)
does not flash
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
이미 열람한 고객
문서
DNA damage and repair mechanism is vital for maintaining DNA integrity. Damage to cellular DNA is involved in mutagenesis, the development of cancer among others.
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.