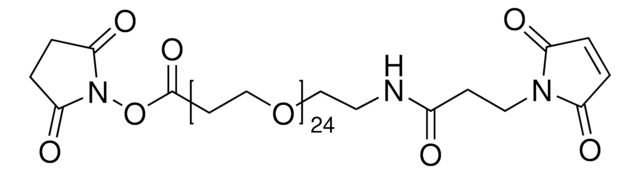
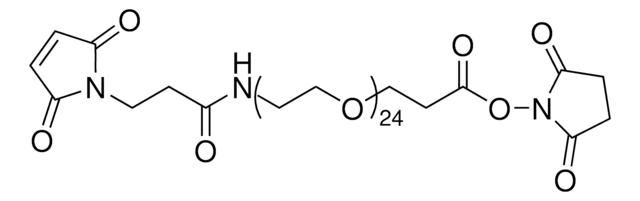
746223
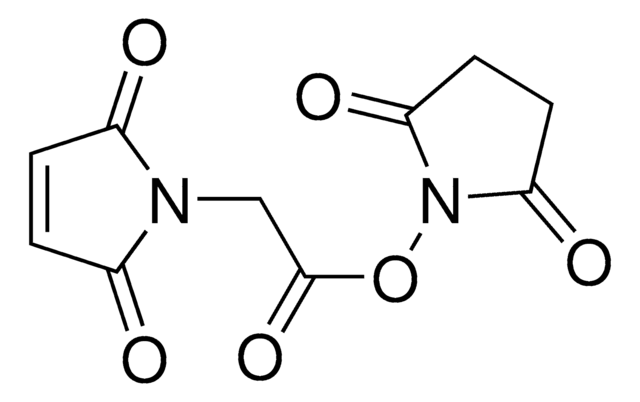
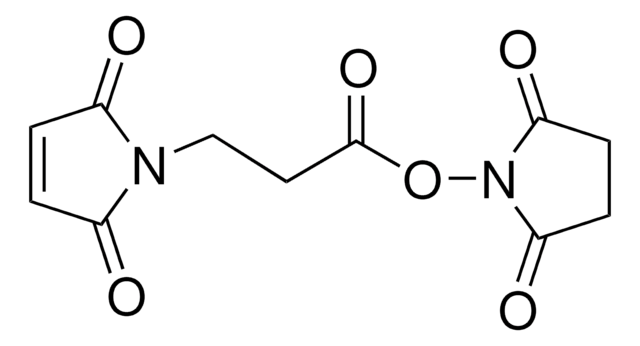
Maleimide-PEG2-succinimidyl ester
≥95%
Synonyme(s) :
3-[2-[2-[[3-(2,5-Dihydro-2,5-dioxo-1H-pyrrol-1-yl)-1-oxopropyl]amino]ethoxy]ethoxy]propanoic acid 2,5-dioxo-1-pyrrolidinyl ester, Maleimide-PEG2-NHS
About This Item
Produits recommandés
Essai
≥95%
Forme
solid
Capacité de réaction
reaction type: Pegylations
Pertinence de la réaction
reagent type: cross-linking reagent
Pf
92-94 °C
Groupe fonctionnel
NHS ester
maleimide
Température de stockage
−20°C
Chaîne SMILES
O=C(N1CCC(NCCOCCOCCC(ON(C(CC2)=O)C2=O)=O)=O)C=CC1=O
InChI
1S/C18H23N3O9/c22-13(5-8-20-14(23)1-2-15(20)24)19-7-10-29-12-11-28-9-6-18(27)30-21-16(25)3-4-17(21)26/h1-2H,3-12H2,(H,19,22)
Clé InChI
TZPDZOJURBVWHS-UHFFFAOYSA-N
Application
Code de la classe de stockage
11 - Combustible Solids
Classe de danger pour l'eau (WGK)
WGK 3
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Faites votre choix parmi les versions les plus récentes :
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Les clients ont également consulté
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique
![O-[N-(3-Maleimidopropionyl)aminoethyl]-O′-[3-(N-succinimidyloxy)-3-oxopropyl]heptacosaethylene glycol ≥90% (oligomer purity)](/deepweb/assets/sigmaaldrich/product/structures/367/864/1c31a65f-501f-4464-8bb3-edff0d131a12/640/1c31a65f-501f-4464-8bb3-edff0d131a12.png)