HWGCRISPR
Sigma Whole Human Genome Lentiviral CRISPR Pool
About This Item
Recommended Products
packaging
pkg of 8x25 μL (vials)
Quality Level
concentration
5x108 VP/ml (via p24 assay)
application(s)
CRISPR
shipped in
dry ice
storage temp.
−70°C
Related Categories
General description
Application
Biochem/physiol Actions
Features and Benefits
- Use CRISPR nucleases to knockout protein-coding genes to assess their function
- Efficiently screen the whole human genome (16,000+ genes) at the bench-top without robotics or specialized equipment
- Increased flexibility consisting of 8 subpools at 23,000 clones per pool (184,000 clones total), from the previous 2 subpools (Gecko v2)
- Expanded gRNA coverage at 10 gRNAs per gene, from the previous 6 gRNAs per gene (Gecko v2)
- Numerous built-in enrichment and depletion controls allows researchers to confidently gauge the success of their pooled screening experiments
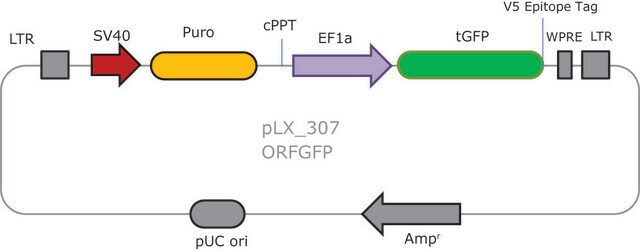
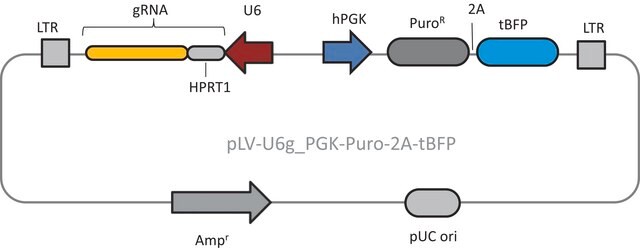
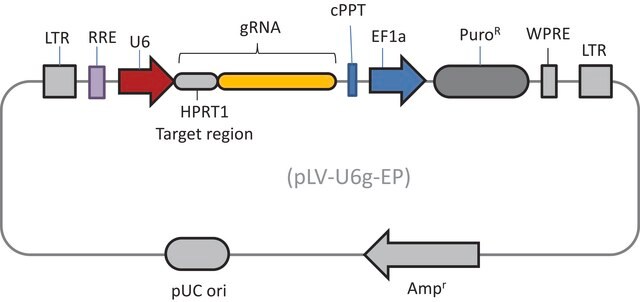
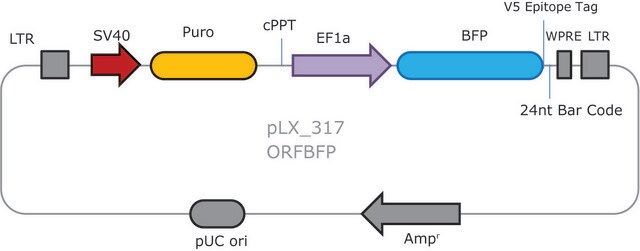
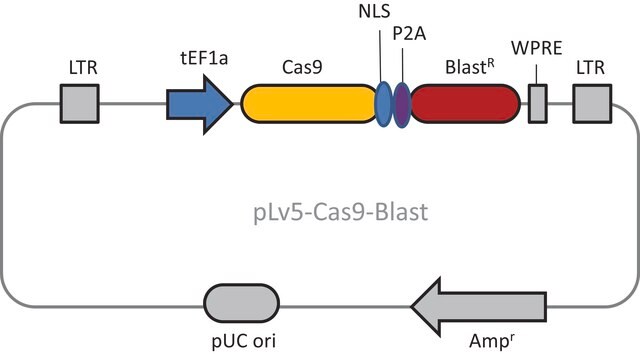
- 2 Vector System (pools are gRNA-only, Cas9 sold separately)
- Ease of optimization: Utilizes same vector system as Gecko v2 allowing for optimization one for both systems
- Minimize off-targeting: stringent gRNA design and tiling rules
Preparation Note
Prior to performing a library-scale screening, two preliminary experiments must be conducted: (1) determine the sensitivity of target cell type to puromycin (kill curve), and (2) determine the functional titer of the lentivirus in your cell type by completing a colony-forming assay (measured in CFU/ml). The calculation of MOI (multiplicity of infection) should be based on the value of CFU. Different cell types vary in transduction efficiency and different lentiviral constructs do not behave identically, so it is critical to optimize your experimental conditions with control lentiviral CRISPR clones (available from Sigma) prior to performing your pooled experiment.
Other Notes
Legal Information
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Our lentiviral vector systems are developed with enhanced safety features. Numerous precautions are in place in the design of our lentiviruses to prevent replication. Good handling practices are a must.
Get tips for handling lentiviruses, optimizing experiment setup, titering lentivirus particles, and selecting helpful products for transduction.
Protocols
You are not alone designing successful CRISPR, RNAi, and ORF experiments. Sigma-Aldrich was the first company to commercially offer lentivirus versions of targeted genome modification technologies and has the expertise and commitment to support new generations of scientists.
FACS (Fluorescence-Activated Cell Sorting) provides a method for sorting a mixed population of cells into two or more groups, one cell at a time, based on the specific light scattering and fluorescence of each cell. This method provides fast, objective, and quantitative recording of fluorescent signals from individual cells.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service