I7634
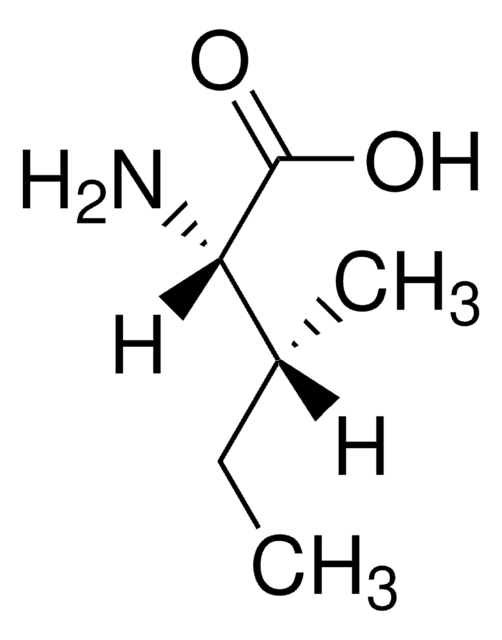
D-Isoleucine
≥98% (TLC)
Sinónimos:
(2R, 3R)-2-Amino-3-methylpentanoic acid
About This Item
Productos recomendados
product name
D-Isoleucine, ≥98% (TLC)
Quality Level
assay
≥98% (TLC)
form
powder
impurities
≤10% allo-isomer
color
white
application(s)
cell analysis
SMILES string
CC[C@@H](C)[C@@H](N)C(O)=O
InChI
1S/C6H13NO2/c1-3-4(2)5(7)6(8)9/h4-5H,3,7H2,1-2H3,(H,8,9)/t4-,5-/m1/s1
InChI key
AGPKZVBTJJNPAG-RFZPGFLSSA-N
¿Está buscando productos similares? Visita Guía de comparación de productos
Categorías relacionadas
Application
- Metabolic Profiling of Urinary Chiral Amino-Containing Biomarkers for Gastric Cancer Using a Sensitive Chiral Chlorine-Labeled Probe by HPLC-MS/MS.: This study by Huang et al. (2021) explores the use of D-Isoleucine as a biomarker for gastric cancer, utilizing advanced metabolic profiling techniques with HPLC-MS/MS to detect chiral amino-containing compounds in urine samples (Huang et al., 2021).
- Isopenicillin N Synthase Binds δ-(L-α-aminoadipoyl)-L-cysteinyl-D-thia-allo-isoleucine Through Both Sulfur Atoms.: Clifton et al. (2011) describe the binding mechanism of Isopenicillin N synthase with a compound involving D-Isoleucine, highlighting its role in antibiotic biosynthesis and enzyme interaction studies (Clifton et al., 2011).
- The Total Structure of the Antibiotic Longicatenamycin.: Shiba and Mukunoki (1975) determined the complete structure of the antibiotic longicatenamycin, which includes D-Isoleucine as a crucial component, contributing to understanding its antibacterial properties (Shiba and Mukunoki, 1975).
Biochem/physiol Actions
Storage Class
11 - Combustible Solids
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Chromatograms
application for HPLCNuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico