PK-RO
Roche
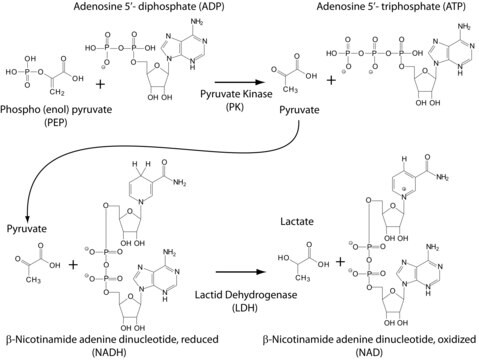
Pyruvate Kinase (PK)
from rabbit muscle
Sinónimos:
PK
About This Item
Productos recomendados
biological source
rabbit muscle
Quality Level
form
suspension
specific activity
~200 units/mg protein (at 25 °C (500 U/mg at 37 °C) with PEP as the substrate.)
packaging
pkg of 1 mL (10128155001 [10 mg])
pkg of 10 mL (10128163001 [100 mg])
manufacturer/tradename
Roche
optimum pH
7.0-7.5
storage temp.
2-8°C
General description
Pyruvate kinase has a molar mass of 237,000 and exists as a tetramer. Each polypeptide chain of this tetramer has a molar mass of 57,200. The enzyme contains two identical catalytic particles called protomers. Each of these protomers contains two polypeptide chains. Each protomer contains one site each for Mn2+ and phosphoenolpyruvate.
Application
Biochem/physiol Actions
Quality
Physical form
Preparation Note
Other Notes
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
No data available
flash_point_c
No data available
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico