11745808910
Roche
Nick Translation Mix
sufficient for 50 labeling reactions, pkg of 200 μL, solution
Sinónimos:
nick translation
About This Item
Productos recomendados
form
solution
Quality Level
usage
sufficient for 50 labeling reactions
packaging
pkg of 200 μL
manufacturer/tradename
Roche
technique(s)
nucleic acid labeling: suitable
color
colorless
pH
~7.5 (68 °F)
solubility
water: miscible
suitability
suitable for fluorescent labeling techniques
suitable for molecular biology
application(s)
genomic analysis
life science and biopharma
storage temp.
−20°C
General description
Individual templates produce consistent results in the standard 90-minutes reaction, and result in an average probe length of 200 base pairs up to 500 base pairs.
Assay Time: 100 minutes
Sample Materials
- Supercoiled and linearized plasmid DNA
- Supercoiled and linearized cosmid DNA
- Purified PCR products
Specificity
Application
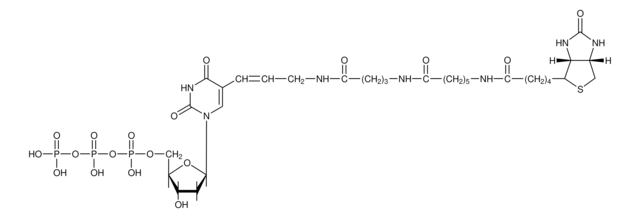
The Nick Translation Mix is designed for direct fluorophore-labeling of in situ probes. Fluorescein-12-dUTP and Tetramethyl-Rhodamine-5-dUTP from Roche Applied Science or other commercially available fluorophor-labeled nucleotides can be combined with the Nick Translation Mix. Direct fluorophore-labeled in situ probes are used for the detection of multi copy or very large hybridization targets on metaphase chromsomes or interphase nuclei.
For a standard labeling reaction using 1 μg template in 20 μl total reaction volume, 4 μl of 5x concentrated fluorophore labeling mix are required.
Components
1 vial with 5x concentrated solution, stabilized reaction buffer in 50% glycerol (v/v), DNA Polymerase I and DNase I.
Quality
Principle
E. coli DNA Polymerase I synthesizes DNA complementary to the intact strand in a 5′?3′ direction using the 3′-OH termini of the nick as a primer. The 5′?3′ exonucleolytic activity of DNA polymerase I simultaneously removes nucleotides in the direction of synthesis. The polymerase activity sequentially replaces the removed nucleotides with isotope-labeled or hapten-labeled deoxyribonucleoside triphosphates. At low temperature (+15°C), the unlabeled DNA in the reaction is thus replaced by newly synthesized labeled DNA.
Storage and Stability
Other Notes
Denaturing of the template before nick translation is not required.
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
does not flash
flash_point_c
does not flash
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico