推薦產品
形狀
ready-to-use solution
品質等級
品質
Protein Mass Spectrometry Calibration Standard
技術
mass spectrometry (MS): suitable
儲存溫度
−20°C
一般說明
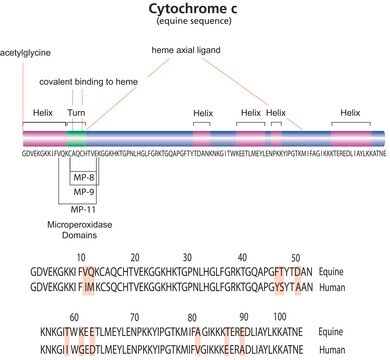
通用蛋白质组学标准(UPS)套件是与生物分子资源设施协会(ABRF)蛋白质组学标准研究小组(sPRG)合作开发的。在一项全面的2005/2006研究中,在ABRF′s sPRG的指导下对这种蛋白质混合物进行了广泛的评估和报告。该研究的结果在ABRF 2006和US HUPO 2006会议上进行了介绍。
應用
通用蛋白质组学标准(UPS)套件预期用于在分析复杂蛋白质样品之前标准化和/或评估质谱(例如LC-MS/MS,MALDI-TOF-MS等)和电泳分析条件。潜在用途包括:
- 将关键实验数据集括起来,以确认分析方法的稳健性
- 使用各种分析策略和仪器在不同实验室中生成的MS或其他蛋白质组学数据的比较
- 识别蛋白质组学分析系统和搜索算法的局限性
- 辅助评价定义不明确样本的数据的外部参考
特點和優勢
为自己发现好处!
- 测试分析策略的功效
- 故障排除和优化您的分析方案
- 在分析关键样品之前确认系统适用性
- 逐日或逐个实验室标准化分析结果
套裝中的組件也可單獨購買
產品號碼
描述
SDS
- T6567Trypsin from porcine pancreas, Proteomics Grade, BioReagent, Dimethylated 20 μgSDS
訊號詞
Danger
危險分類
Acute Tox. 4 Oral - Eye Dam. 1 - Repr. 1B - Resp. Sens. 1 - Skin Irrit. 2 - STOT SE 3
標靶器官
Respiratory system
儲存類別代碼
6.1C - Combustible acute toxic Cat.3 / toxic compounds or compounds which causing chronic effects
水污染物質分類(WGK)
WGK 3
客戶也查看了
David L Tabb et al.
Journal of proteome research, 9(2), 761-776 (2009-11-20)
The complexity of proteomic instrumentation for LC-MS/MS introduces many possible sources of variability. Data-dependent sampling of peptides constitutes a stochastic element at the heart of discovery proteomics. Although this variation impacts the identification of peptides, proteomic identifications are far from
Stephane Houel et al.
Journal of proteome research, 9(8), 4152-4160 (2010-06-29)
A complicating factor for protein identification within complex mixtures by LC/MS/MS is the problem of "chimera" spectra, where two or more precursor ions with similar mass and retention time are co-sequenced by MS/MS. Chimera spectra show reduced scores due to
Matthew The et al.
Nature communications, 11(1), 3234-3234 (2020-06-28)
In shotgun proteomics, the analysis of label-free quantification experiments is typically limited by the identification rate and the noise level in the quantitative data. This generally causes a low sensitivity in differential expression analysis. Here, we propose a quantification-first approach
Christian J Koehler et al.
Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry, 25(1), 61-66 (2019-11-02)
Proteolytic digestion prior to LC-MS analysis is a key step for the identification of proteins. Digestion of proteins is typically performed with trypsin, but certain proteins or important protein sequence regions might be missed using this endoproteinase. Only few alternative
Amanda G Paulovich et al.
Molecular & cellular proteomics : MCP, 9(2), 242-254 (2009-10-28)
Optimal performance of LC-MS/MS platforms is critical to generating high quality proteomics data. Although individual laboratories have developed quality control samples, there is no widely available performance standard of biological complexity (and associated reference data sets) for benchmarking of platform
相關內容
Standardize Your Research. We offer both the Universal Proteomics Standard and the Proteomics Dynamic range Standard as complex, well-defined, well characterized reference standards for mass spectrometry.
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務