KMM-201NV
KOD One™ PCR Master Mix -BLUE-
Ready-to-use 2X hot-start PCR master mix with a modified KOD DNA polymerase optimized for ultra-fast and convenient high-fidelity PCR
同義詞:
Fast Hot-start PCR, High-fidelity DNA polymerase, KOD One™ PCR Ready Mix
登入查看組織和合約定價
全部照片(2)
About This Item
分類程式碼代碼:
41106314
NACRES:
NA.55
推薦產品
生物源
Escherichia coli (carrying the cloned UKOD DNA polymerase gene)
品質等級
形狀
liquid
用途
sufficient for 25 reactions
sufficient for 500 reactions
特點
Fast PCR
High Fidelity PCR
dNTPs included
hotstart
技術
PCR: suitable
輸入
crude DNA
運輸包裝
dry ice
儲存溫度
−20°C
一般說明
KOD One™ PCR master Mix -Blue- is a ready-to-use 2 x PCR master mix containing a novel genetically modified KOD DNA polymerase (UKOD) along with a new elongation accelerator, enabling fast PCR with an extension time of 5 sec/ kb for template DNA <10kb. This master mix has greater efficiency and flexibility than conventional PCR enzymes with an ability to amplify from crude specimens such as blood and mouse cell lysates. In addition to crude samples, it can be used on templates containing uracils (dU). The master mix can use primers containing both inosines (dI) and dU for amplification.
This hot start KOD polymerase is antibody-inactivated with two types of antibodies that prevent the 5′→3′ polymerase activity and 3′→5′ exonuclease activity. Upon heat-activation, the 3′→5′ proof-reading ability of the enzyme generates blunt-end PCR products.
This hot start KOD polymerase is antibody-inactivated with two types of antibodies that prevent the 5′→3′ polymerase activity and 3′→5′ exonuclease activity. Upon heat-activation, the 3′→5′ proof-reading ability of the enzyme generates blunt-end PCR products.
應用
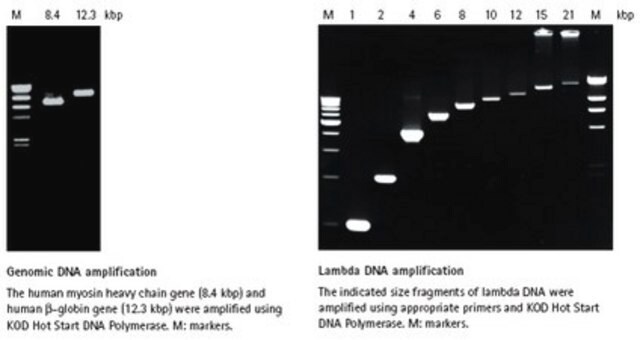
- Genomic DNA amplification
- cDNA amplification
- Direct PCR
- Colony PCR
- Amplification of NGS libraries
- Site direct gene mutation
Compatible Sample Types: Serum, Plasma, Cell, Plant, soil extraction, DNA, nail, hair etc.
Sample Volume Needed: about 50ng DNA
特點和優勢
- Fast: Amplifies the targets using the following very short conditions:
o 1∼ 10 kb: 5 sec/ kb
o 10 kb∼: 10 sec/ kb
- Ready-to-use: 2X premixed solution of UKOD DNA polymerase, buffer, dNTPs, and MgCl2. KOD One PCR Master Mix Blue includes a loading dye (BPB) to allow direct loading onto agarose gels.
- High Fidelity: Approximately 80-fold higher fidelity than conventional Taq DNA polymerase. This mix can be used for various purposes where this would be an advantage (e.g., in the preparation of long target amplicons for sequencing).
- High Efficiency: Enables high throughput PCR and ultra-fast PCR cycling conditions with an extension time of as quick as 5 sec/kb.
- Crude Sample PCR: Effective for direct PCR amplification of crude samples such as biological samples, food samples, soil extract, etc.
- High flexibility: Amplifies templates containing uracils (dU). Also, amplifies fragments using degenerate primers with inosines (dI) and uracils (dU)
包裝
KMM-201NV - 5X1ML is sufficient for 500 reactions at 20 μL each or for 200 reactions at 50 μL each.
KMM-201NVS - 0.25ML is sufficient for 25 reactions at 20 μL each or for 10 reactions at 50 μL each.
KMM-201NVS - 0.25ML is sufficient for 25 reactions at 20 μL each or for 10 reactions at 50 μL each.
儲存和穩定性
Stored at -20 °C for a year. 4 °C for a month.
其他說明
For R&D use only. Not for drug, household, or other uses.
法律資訊
*Manufactured by Toyobo and distributed by Sigma-Aldrich. Not available in Japan.
KOD One is a trademark of Toyobo Co., Ltd.
訊號詞
Warning
危險聲明
防範說明
危險分類
Met. Corr. 1
儲存類別代碼
8B - Non-combustible corrosive hazardous materials
水污染物質分類(WGK)
WGK 2
閃點(°F)
Not applicable
閃點(°C)
Not applicable
Moe Yokoshi et al.
Molecular cell, 78(2), 224-235 (2020-02-29)
Formation of self-associating loop domains is a fundamental organizational feature of metazoan genomes. Here, we employed quantitative live-imaging methods to visualize impacts of higher-order chromosome topology on enhancer-promoter communication in developing Drosophila embryos. Evidence is provided that distal enhancers effectively
Nuria Cortes-Silva et al.
Current biology : CB, 30(4), 561-572 (2020-02-08)
Accurate chromosome segregation requires assembly of the multiprotein kinetochore complex at centromeres. In most eukaryotes, kinetochore assembly is primed by the histone H3 variant CenH3 (also called CENP-A), which physically interacts with components of the inner kinetochore constitutive centromere-associated network
相關內容
KOD One™ PCR Master Mix overview for ultra-fast PCR with high specificity, fidelity, and yield
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務