推薦產品
生物源
bacterial (Brevibacterium linens)
品質等級
形狀
solution
比活性
10000 U/mL
包裝
pkg of 1,000 U (11558170001 [10 U/μl])
pkg of 200 U (11558161001 [10 U/μl])
製造商/商標名
Roche
參數
37 °C optimum reaction temp.
顏色
colorless
pH值
8.1 (39 °F)
溶解度
water: miscible
適合性
suitable for molecular biology
應用
life science and biopharma
sample preparation
異物活動
Endonucleases, none detected (up to 20 U with MWM II-DNA)
Endonucleases, none detected (up to 20U with pBR 322-DNA)
運輸包裝
dry ice
儲存溫度
−20°C
一般說明
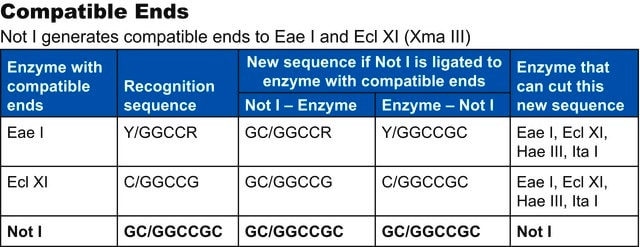
Compatible ends
Bln I ends are compatible with ends generated by Nhe I, Spe I and Xba I.
Isoschizomers
Bln I is an isoschizomer of Avr II.
Note: The complete 13 site Avr II restriction map of the E.coli genome has been reported.
Methylation sensitivity
The enzyme is not known to be affected by methylation.
特異性
CCTAGG
Restriction site: C↓CTAGG
C↓CTAGG
Heat inactivation: No inactivation of Bln I after incubation at 65 °C for 15 minutes.
品質
1 μg λDNA is incubated for 16 hours in 50 μl SuRE/Cut Buffer H with an excess of Bln I. The number of enzyme units which do not change the enzyme-specific pattern is stated in the certificate of analysis.
Absence of exonuclease activity
Approximately 5 μg [3H] labeled calf thymus DNA are incubated with 3 μl Bln I for 4 hours at +37°C in a total volume of 100 μl 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithioerythritol, pH approximately 7.5. Under these conditions, no release of radioactivity is detectable, as stated in the certificate of analysis.
Typical ligation and recutting assay
Bln I fragments obtained by complete digestion of 1 μg λ × EcoR I DNA ligated for 16 hours at +4°C with 1 U T4 DNA Ligase in 10 μl buffer that contains 66 mM Tris-HCl, 5 mM MgCl2, 5 mM Dithiothreitol, 1 mM ATP, pH 7.5 (at +20°C). The percentages of product that can be ligated and subsequently recut with Bln I and EcoR I (yielding the typical pattern of λ × EcoR I × Bln I fragments) are stated under "Lig" and "Rec" in the certificate of analysis.
DNA分析
- λ: 2
- φX174: 0
- Ad2: 2
- M13mp7: 0
- M13mp18:0
- pBR322: 0
- pBR328: 0
- pUC18: 0
- SV40: 2
單位定義
儲存和穩定性
分析報告
Bln I has been tested in Pulsed-Field Gel Electrophoresis (on bacterial chromosomes). For cleavage of genomic DNA (E.coli C 600) embedded in agarose for PFGE analysis, we recommend using 10 U of enzyme/μg DNA and 4 hour incubation.
The buffer in bold is recommended for optimal activity
- A: 25-50%
- B: 50-75%
- H: 100%
- L: 0-10%
- M: 25-50%
其他說明
僅套裝組件
- Enzyme Solution
- SuRE/Cut Buffer H 10x concentrated
儲存類別代碼
12 - Non Combustible Liquids
水污染物質分類(WGK)
WGK 1
閃點(°F)
does not flash
閃點(°C)
does not flash
客戶也查看了
文章
The term “Restriction enzyme” originated from the studies of Enterobacteria phage λ (lambda phage) in the laboratories of Werner Arber and Matthew Meselson.
The term “Restriction enzyme” originated from the studies of Enterobacteria phage λ (lambda phage) in the laboratories of Werner Arber and Matthew Meselson.
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務