04-1488
抗-HIRA抗体,克隆WC119
clone WC119, from mouse
同義詞:
DiGeorge critical region gene 1, HIR (histone cell cycle regulation defective) homolog A (S. cerevisiae), HIR histone cell cycle regulation defective homolog A, HIR histone cell cycle regulation defective homolog A (S. cerevisiae), TUP1-like enhancer of
登入查看組織和合約定價
全部照片(1)
About This Item
分類程式碼代碼:
12352203
eCl@ss:
32160702
NACRES:
NA.41
推薦產品
生物源
mouse
品質等級
抗體表格
purified immunoglobulin
抗體產品種類
primary antibodies
無性繁殖
WC119, monoclonal
物種活性
mouse
物種活性(以同源性預測)
human (based on 100% sequence homology)
技術
immunoprecipitation (IP): suitable
western blot: suitable
同型
IgG1κ
NCBI登錄號
UniProt登錄號
運輸包裝
wet ice
目標翻譯後修改
unmodified
基因資訊
human ... HIRA(7290)
一般說明
HIRA(分裂蛋白1的TUP1样增强子)是一种主要位于细胞核中的蛋白。它充当组蛋白chaparone,优先将组蛋白H3.3置于核小体上。在多种组织中广泛表达,并在胚胎发生过程中表达。 数据表明,HIRA在转录调控机制中起着与酵母HIR1和HIR2共同作用相似的作用。已显示HIRA蛋白与HIRIP3,HIRIP5,组蛋白H2B和H4以及PAX3相互作用。HIRA可能在DiGeorge综合征(DGS)(一种发育障碍)的病因中起作用。该疾病的临床特征包括胸腺和甲状旁腺的缺失或发育不全,心血管畸形,面部发育不良,腭裂和智力低下。
特異性
该抗体可识别HIRA。
免疫原
标记GST的重组蛋白,对应于人HIRA。
表位:未知
應用
免疫沉淀分析: 先前批次已在一个独立实验室中进行了IP测试。(Hall, C., et al. (2001).Molecular and Cellular Biology.21(5):1854-1865.)
抗HIRA抗体,克隆WC119是用于检测HIRA(也称为HIR(组蛋白细胞周期调节缺陷)同源物)的小鼠单克隆抗体,&已在WB & IP中进行了验证。
研究子类别
染色质生物学
Chaperones
染色质生物学
Chaperones
研究类别
表观遗传学&核功能
表观遗传学&核功能
品質
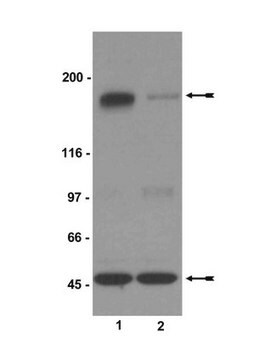
通过蛋白质印迹在NIH/3T3细胞裂解液中进行了评估。
蛋白质印迹分析:0.44 µg/mL抗体可在10 µg NIH/3T3细胞裂解液中检测到HIRA。
蛋白质印迹分析:0.44 µg/mL抗体可在10 µg NIH/3T3细胞裂解液中检测到HIRA。
標靶描述
~112 kDa
外觀
形式:纯化
纯化的小鼠单克隆IgG1κ溶于含0.1 M Tris-甘氨酸(pH 7.4),150 mM NaCl和0.05%叠氮化钠的缓冲液中。
蛋白G纯化
儲存和穩定性
自收到之日起,在2-8°C条件下可稳定保存1年。
分析報告
对照
NIH/3T3细胞裂解液
NIH/3T3细胞裂解液
其他說明
浓度:请参考批次特异性浓缩物的分析证书。
免責聲明
除非我们的产品目录或产品附带的其他公司文档另有说明,否则我们的产品仅供研究使用,不得用于任何其他目的,包括但不限于未经授权的商业用途、体外诊断用途、离体或体内治疗用途或任何类型的消费或应用于人类或动物。
未找到適合的產品?
試用我們的產品選擇工具.
儲存類別代碼
12 - Non Combustible Liquids
水污染物質分類(WGK)
WGK 1
閃點(°F)
Not applicable
閃點(°C)
Not applicable
分析證明 (COA)
輸入產品批次/批號來搜索 分析證明 (COA)。在產品’s標籤上找到批次和批號,寫有 ‘Lot’或‘Batch’.。
Tetsuro Komatsu et al.
Journal of virology, 86(12), 6701-6711 (2012-04-13)
In infected cells, the chromatin structure of the adenovirus genome DNA plays critical roles in its genome functions. Previously, we reported that in early phases of infection, incoming viral DNA is associated with both viral core protein VII and cellular
Zhexin Zhu et al.
Cell stem cell, 20(2), 274-289 (2016-12-13)
The chromatin landscape and cellular metabolism both contribute to cell fate determination, but their interplay remains poorly understood. Using genome-wide siRNA screening, we have identified prohibitin (PHB) as an essential factor in self-renewal of human embryonic stem cells (hESCs). Mechanistically, PHB
Regional identity of human neural stem cells determines oncogenic responses to histone H3.3 mutants.
Raul Bardini Bressan et al.
Cell stem cell, 28(5), 877-893 (2021-02-26)
Point mutations within the histone H3.3 are frequent in aggressive childhood brain tumors known as pediatric high-grade gliomas (pHGGs). Intriguingly, distinct mutations arise in discrete anatomical regions: H3.3-G34R within the forebrain and H3.3-K27M preferentially within the hindbrain. The reasons for
Kristina Ivanauskiene et al.
Genome research, 24(10), 1584-1594 (2014-07-23)
Histone variant H3.3 is deposited in chromatin at active sites, telomeres, and pericentric heterochromatin by distinct chaperones, but the mechanisms of regulation and coordination of chaperone-mediated H3.3 loading remain largely unknown. We show here that the chromatin-associated oncoprotein DEK regulates
Camille Cohen et al.
PLoS pathogens, 14(9), e1007313-e1007313 (2018-09-21)
Herpes simplex virus 1 (HSV-1) latency establishment is tightly controlled by promyelocytic leukemia (PML) nuclear bodies (NBs) (or ND10), although their exact contribution is still elusive. A hallmark of HSV-1 latency is the interaction between latent viral genomes and PML
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務