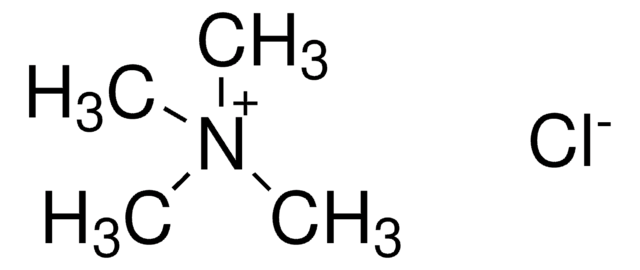
T3411
Tetramethylammonium chloride solution
for molecular biology
Synonym(s):
N,N,N-Trimethylmethanaminium chloride
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
CAS Number:
MDL number:
UNSPSC Code:
12352107
PubChem Substance ID:
NACRES:
NA.31
Recommended Products
grade
for molecular biology
Quality Level
concentration
5 M
foreign activity
DNase, RNase, none detected
SMILES string
[Cl-].C[N+](C)(C)C
InChI
1S/C4H12N.ClH/c1-5(2,3)4;/h1-4H3;1H/q+1;/p-1
InChI key
OKIZCWYLBDKLSU-UHFFFAOYSA-M
General description
Tetramethylammonium binds AT-rich DNA polymers while concomitantly abolishing the preferential melting of AT versus GC base pairs. It is supplied as a 0.2 μm filtered solution in 18 megohm water.
Application
Tetramethylammonium chloride solution (TMAC) has been used:
- in the preparation of hybridization cocktail for array hybridization and scanning
- in next-generation sequencing (NGS), and genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) library preparation
- in the preparation of TMAC buffer and bead hybridization mixture for hybridization and detection
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
W B Melchior et al.
Proceedings of the National Academy of Sciences of the United States of America, 70(2), 298-302 (1973-02-01)
Several small alkylammonium ions can eliminate, or even reverse, the usual dependence of the DNA transition temperature on base composition. For example, in 3 M tetramethylammonium chloride, or 2.4 M tetraethylammonium chloride, DNAs of different base compositions all melt at
Nikolay L Malinin et al.
Nature protocols, 16(12), 5592-5615 (2021-11-14)
Genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) is a sensitive, unbiased, genome-wide method for defining the activity of genome-editing nucleases in living cells. GUIDE-seq is based on the principle of efficient integration of an end-protected double-stranded oligodeoxynucleotide
Hybridization of genomic DNA to oligonucleotide probes in the presence of tetramethylammonium chloride.
A G DiLella et al.
Methods in enzymology, 152, 447-451 (1987-01-01)
Xiao-Yong Li et al.
Methods in molecular biology (Clifton, N.J.), 809, 3-26 (2011-11-25)
Immunoprecipitation of cross-linked chromatin in combination with microarrays (ChIP-chip) or ultra high-throughput sequencing (ChIP-seq) is widely used to map genome-wide in vivo transcription factor binding. Both methods employ initial steps of in vivo cross-linking, chromatin isolation, DNA fragmentation, and immunoprecipitation.
Markus Welcker et al.
Genes & development, 27(23), 2531-2536 (2013-12-04)
The Fbw7 tumor suppressor targets a broad network of proteins for ubiquitylation. Here we show critical functions for Fbw7 dimerization in regulating the specificity and robustness of degradation. Dimerization enables Fbw7 to target substrates through concerted binding to two suboptimal
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service