HSAMPURO
Human Whole Genome SAM CRISPRa Pooled Lentiviral Library Kit Puro
Synonym(s):
Human CRISPRa Library, SAM CRISPRa Library
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
41106621
NACRES:
NA.51
Recommended Products
packaging
pkg of 40 vials (8x25μL aliquots for each of the 5 kit components)
application(s)
CRISPR
shipped in
dry ice
storage temp.
−70°C
General description
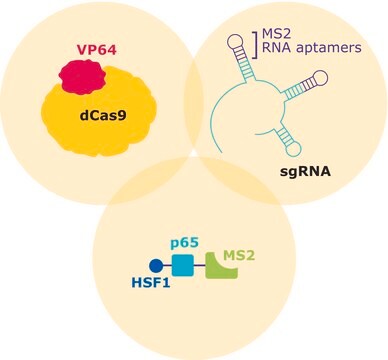
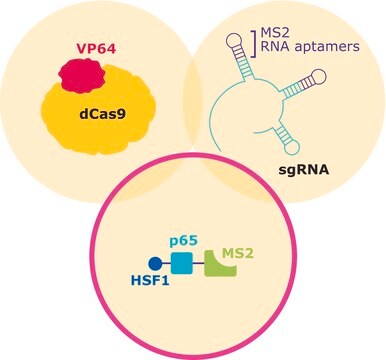
The transcriptional activator VP64 fused to nuclease-dead SpCas9 (dCas9), forms a ribonucleoprotein complex (RNP) with the CRISPR guide RNA (gRNA). The stem- and tetra-loop sequences in the gRNA scaffold have been modified into minimal hairpin RNA aptamers, which selectively bind dimerized MS2 bacteriophage coat proteins. MS2 coat protein is fused to the p65 subunit of NF-kappaB (NFκB) and the activation domain of human heat-shock factor 1 (HSF1). The guide RNA contains two aptamers, each capable of binding two MS2 coactivator proteins, effectively recruiting four co-activators for every CRISPR targeting activator complex.
The dCas9-VP64,MS2-p65-HSF1 helper constructs and gRNA are delivered as three separate plasmids/Lentiviral particles. In contrast to arrayed gRNA libraries containing each clone in a separate plate well, pooled gRNA libraries contain thousands of individual gRNAs in a single tube enabling efficient screening of the whole human genome (16,000+ genes) at the bench-top without robotics or specialized equipment. The SAM CRISPR library is provided in 3 sub pools. Each sub-pool contains approximately 23,500 gRNAs, with 3 gRNAs per RefSeqID. The whole genome library contains approximately 70,000 gRNAs, in total, targeting 19,000 unique gene symbols (including alternative isoforms encompassing 23,500 RefSeqIDs).
100 non-targeting (i.e. negative) control gRNAs are included within each pool for use as a baseline in the statistical characterization of changes in gRNA frequency as measured by deep sequencing. Negative control gRNAs share minimal homology with the target genome and should not undergo significant changes in representation throughout control cell treatments. Due to the nature of CRISPR activator screening experiments, positive controls must be determined empirically, taking into account the specific phenotype to be explored in the screen. Together, these controls create many options for upfront experimental design and downstream data analysis.
The dCas9-VP64,MS2-p65-HSF1 helper constructs and gRNA are delivered as three separate plasmids/Lentiviral particles. In contrast to arrayed gRNA libraries containing each clone in a separate plate well, pooled gRNA libraries contain thousands of individual gRNAs in a single tube enabling efficient screening of the whole human genome (16,000+ genes) at the bench-top without robotics or specialized equipment. The SAM CRISPR library is provided in 3 sub pools. Each sub-pool contains approximately 23,500 gRNAs, with 3 gRNAs per RefSeqID. The whole genome library contains approximately 70,000 gRNAs, in total, targeting 19,000 unique gene symbols (including alternative isoforms encompassing 23,500 RefSeqIDs).
100 non-targeting (i.e. negative) control gRNAs are included within each pool for use as a baseline in the statistical characterization of changes in gRNA frequency as measured by deep sequencing. Negative control gRNAs share minimal homology with the target genome and should not undergo significant changes in representation throughout control cell treatments. Due to the nature of CRISPR activator screening experiments, positive controls must be determined empirically, taking into account the specific phenotype to be explored in the screen. Together, these controls create many options for upfront experimental design and downstream data analysis.
Features and Benefits
- Highly specific and highly active
- Sequence verified high purity, high titer lentiviral particles
- Activates genes through transcriptional activation rather than cDNA based overexpression
Learn more about SAM CRISPR Activators at SigmaAldrich.com/CRISPRa
Components
This kit contains 5 components:
2 Helper Constructs with a minimum concentration of 1x106 VP/mL(via p24 assay)
3 Subpools with a minimum conecentration of 5x108 VP/mL(via p24 assay)
2 Helper Constructs with a minimum concentration of 1x106 VP/mL(via p24 assay)
- 8 x 25μl vials of dCas9-VP64-Blasticidin SAM CRISPRa Helper Construct 1 Lentiviral Transduction Particles
- 8 x 25μl vials of MS2-P65-HSF1-Hygromycin SAM CRISPRa Helper Construct 2 Lentiviral Transduction Particles
3 Subpools with a minimum conecentration of 5x108 VP/mL(via p24 assay)
- 8 x 25μl vials of Human SAM CRISPRa Library Pool 1 - Puro
- 8 x 25μl vials of Human SAM CRISPRa Library Pool 2 - Puro
- 8 x 25μl vials of Human SAM CRISPRa Library Pool 3 - Puro
Principle
CRISPR/Cas systems are employed by bacteria and archaea as a defense against invading viruses and plasmids. Recently, the type II CRISPR/Cas system from the bacterium Streptococcus pyogenes has been engineered to function in eukaryotic systems using two molecular components: a single Cas9 protein and a non-coding guide RNA (gRNA). The Cas9 endonuclease can be rendered inactive (dCas9) with mutations to the two protein domains, RuvC and HnH (D10A and H840A respectively), which are responsible for nuclease activity. The nuclease deficient protein can then be fused with the transcriptional activator VP64 and used in conjunction with a guide RNA modified with MS2 RNA aptamers that function to recruit the additional transcriptional coactivators p65 and HSF1. The assembled SAM complex is then used as a cargo delivery system to target gene promoters, enabling site-specific transcriptional activation of the gene of interest.
Unit Definition
VP/ml is the concentration unit of measure for viral titer estimated by p24 assay.
Storage Class Code
10 - Combustible liquids
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Sorry, we don't have COAs for this product available online at this time.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
Konermann S, et al
Nature, 517, 583-588 (2015)
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
Konermann S, et al.
Nature, 517, 583-588 (2015)
Genome-scale CRISPR-Cas9 Knockout and Transcriptional Activation Screening.
Joung, S. et al.
Nature Protocols, 12, 828-863 (2017)
Julia Joung et al.
Nature protocols, 12(4), 828-863 (2017-03-24)
Forward genetic screens are powerful tools for the unbiased discovery and functional characterization of specific genetic elements associated with a phenotype of interest. Recently, the RNA-guided endonuclease Cas9 from the microbial CRISPR (clustered regularly interspaced short palindromic repeats) immune system
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service