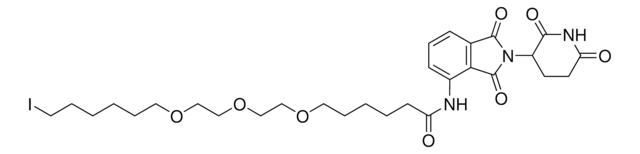
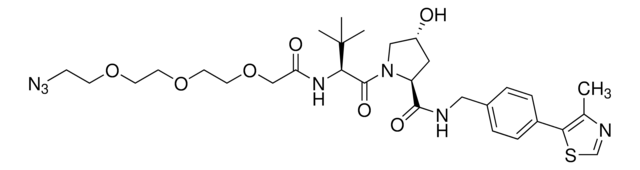
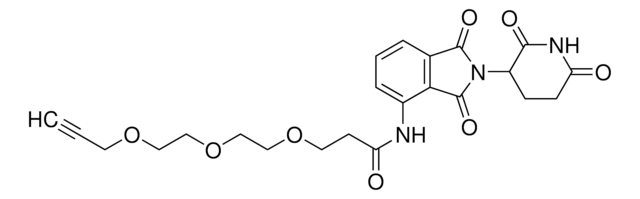
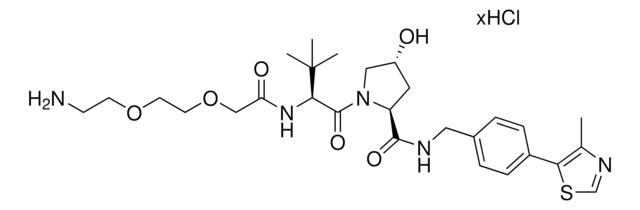
901503
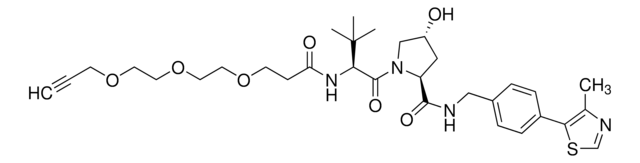
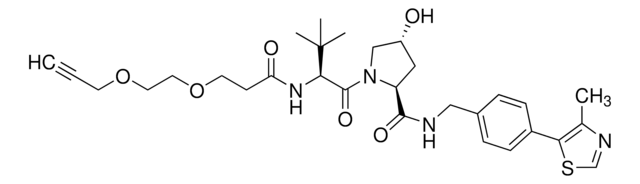
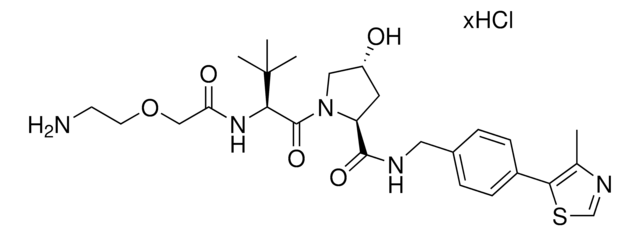
(S,R,S)-AHPC-PEG1-Alkyne
≥95%
别名:
(2S,4R)-1-((S)-3,3-Dimethyl-2-(3-(prop-2-yn-1-yloxy)propanamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide, CrosslinkerE3 Ligase ligand conjugate, Protein degrader building block for PROTAC® research, Template for synthesis of targeted protein degrader, VH032 conjugate
About This Item
推荐产品
品質等級
化驗
≥95%
形狀
powder or crystals
反應適用性
reaction type: click chemistry
reagent type: ligand-linker conjugate
官能基
alkyne
儲存溫度
2-8°C
SMILES 字串
O=C(NCC1=CC=C(C2=C(C)N=CS2)C=C1)[C@H](C[C@@H](O)C3)N3C([C@H](C(C)(C)C)NC(CCOCC#C)=O)=O
應用
Automate your VHL-PEG based PROTACs with Synple Automated Synthesis Platform (SYNPLE-SC002)
其他說明
Portal: Building PROTAC® Degraders for Targeted Protein Degradation
Targeted Protein Degradation by Small Molecules
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Impact of linker length on the activity of PROTACs
法律資訊
儲存類別代碼
11 - Combustible Solids
水污染物質分類(WGK)
WGK 3
閃點(°F)
Not applicable
閃點(°C)
Not applicable
其他客户在看
商品
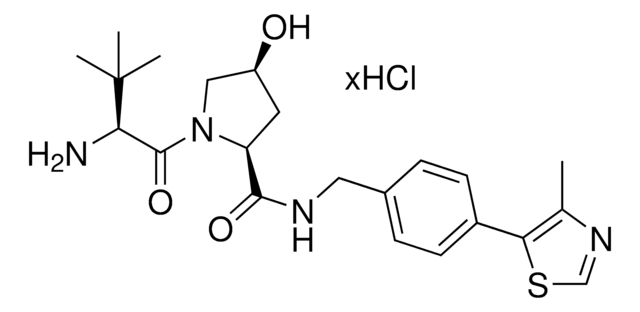
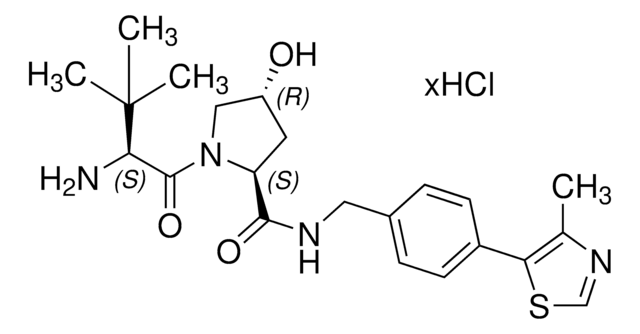
Partial PROTACs are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Partial PROTACs are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Partial PROTACs are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Partial PROTACs are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门