WGA2
GenomePlex® Complete Whole Genome Amplification (WGA) Kit
Optimized kit with enzyme for amplifying a variety of DNA including FFPE tissue
Synonym(s):
Whole genome amplification kit
About This Item
Recommended Products
Related Categories
General description
Application
- microarray analysis
- SNP analysis
- STR analysis
- DNA archiving
Features and Benefits
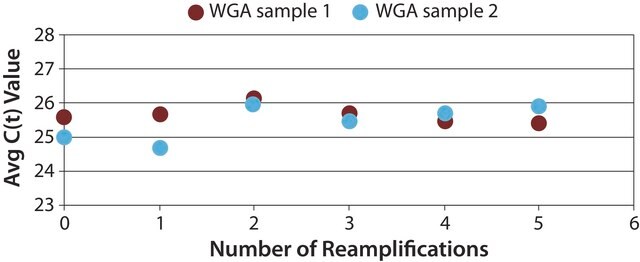
- Higher yield from minimal template: Amplification of nanogram amounts of genomic DNA to microgram yields (around 10 μg) in less than about three hoursrs
- Nanograms of samples can be preserved at –20 °C for future use
- Choose from a variety of DNA sources: whole blood, buccal swab, blood card, plant, soil, & formalin-fixed paraffin-embedded tissue (FFPE)
- Whole-genome amplification (WGA) DNA polymerase increases the amplification accuracy
- Whole-genome representation with no detectable allele bias
- Compatible with many downstream applications such as TaqMan® assays, single nucleotide polymorphism (SNP) analysis, comparative genomic hybridization (CGH) analysis
Other Notes
Legal Information
Kit Components Also Available Separately
- W4502Water, Nuclease-Free Water, for Molecular BiologySDS
related product
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Resp. Sens. 1
Storage Class Code
12 - Non Combustible Liquids
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
In recent years, array-based Comparative Genomic Hybridization (aCGH) has been refined to determine chromosomal changes at progressively higher resolutions. This evolving technology is, however, hampered by the large DNA input requirement—a minimum of 150,000 copies of a human genome, or 0.5 μg, are generally needed per sample to rocess one CGH array.
In recent years, array-based Comparative Genomic Hybridization (aCGH) has been refined to determine chromosomal changes at progressively higher resolutions. This evolving technology is, however, somewhat hampered by the large DNA input requirement—a minimum of 150,000 copies of a human genome, or 0.5 μg, are generally needed per sample to process one CGH array.
The assessment of DNA quality is a crucial first step in acquiring meaningful data from formalin-fixed paraffin-embedded (FFPE) tissues, and other sources of damaged DNA. Using intact genomic DNA is key for successful analysis of chromosomal aberrations (e.g. SNP analysis, LOH, aCGH, etc.).
Protocols
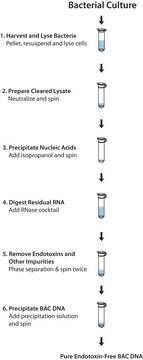
Genomic DNA from soil samples can be easily damaged by nucleases and contaminating debris resulting in low DNA yields. As a result, the researcher’s ability to perform downstream analysis may be compromised. After isolating DNA from the soil sample, the GenomePlex® Whole Genome Amplification Protocol is followed
Whole genome amplification (WGA) of plasma and serum DNA presents a unique challenge due to the small amount of nucleic acid in such samples.
This protocol provides a simple and convenient method to isolate, amplify and purify genomic DNA from saliva
Blood cards provide the convenience of archiving small volumes of blood. However, many times genomic DNA from these samples is limited, This protocol provides a simple and convenient method to extract genomic DNA from a blood card. Once the DNA has been extracted, it can then be amplified using the amplification protocol
Related Content
GenomePlex® Whole Genome Amplification is the method of extracting DNA from the animal sample. GenomePlex® products have been used to amplify genomic DNA from chicken, porcine, bovine, fish, and shrimp source.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service