Qualitative multiplex PCR assay for assessing DNA quality from FFPE tissues and other sources of damaged DNA
Steve Michalik, Christopher William
The assessment of DNA quality is a crucial first step in acquiring meaningful data from formalin-fixed paraffin-embedded (FFPE) tissues, and other sources of damaged DNA. Using intact genomic DNA is key for successful analysis of chromosomal aberrations (e.g. SNP analysis, LOH, aCGH, etc.). FFPE tissues are increasingly being used as a source of DNA for molecular characterization of chromosomal aberrations, but it is well known that formalin reacts with nucleic acids to cause irreversible damage resulting in DNA samples of poor quality that may not work in downstream processes.1,2 There are multiple factors that contribute to DNA damage within these sample collections, namely fixation conditions and the age of the sample. As a result, it is difficult to determine the extent of damage from sample to sample. To address this issue, We have developed a simple, qualitative, gel-based multiplex PCR assay that can be used to determine DNA quality before performing tedious and expensive downstream processes such as aCGH. Multiplex PCR using primer sets that amplify fragments of increasing size have been demonstrated to be an effective quality control tool for identifying overly damaged samples. The ability to amplify the larger PCR products is predictive of high quality DNA that is capable of successful molecular manipulation, including WGA.3,4
Material and Methods
The protocol for the gel-based multiplex PCR assay for predicting DNA degradation is provided below. The assay consists of five primer sets, derived from the NCBI UniSTS database, that amplify products ranging from 132 bp to 295 bp (Table 1).
NCBI UniSTS database
Some, or all, of these products will fail to amplify as DNA sample quality fades, allowing the classification of genomic DNA quality based on the number and size of fragments amplified, with high quality genomic DNA producing all five amplicons. This assay cannot predict the usefulness of damaged DNA without prior validation work. For instance, a sample may be too fragmented to perform aCGH, but may still be useful in qPCR-based SNP analysis. By performing correlative experiments to empirically determine a quality threshold, one can ensure the repeatability of their downstream experiments. For example, some applications may require that all five amplicons be produced in order to predict a successful outcome, whereas others, such as qPCR, may only require the presence of a single band (Figure 1). In this way, one can compare samples from different sources and repeatability of experiments is ensured.
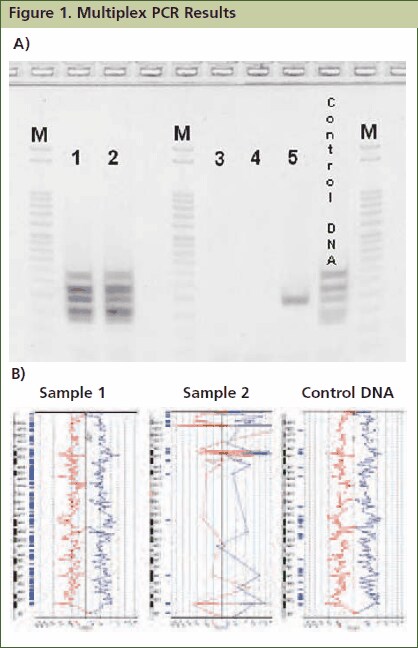
Figure 1.Approximately 1mg of tissue was collected from five FFPE tissue samples followed by processing with the GenomePlex Tissue Whole Genome Amplification Kit (Product No. WGA2) as outlined in the technical bulletin. (A) 5 μL of undiluted WGA2 tissue lysate was subjected to multiplex PCR amplification as outlined below, and 5 μL of each reaction was resolved on a 4% agarose gel (Invitrogen # G6000-04). The 100 bp DNA ladder (Invitrogen catalog # 15628-019) was used as a size standard. All five bands were amplified in lanes 1 and 2 indicating that these FFPE tissue lysates contain high quality genomic DNA, whereas lanes 3, 4 and 5 contain low quality DNA since all, or most, of the multiplex PCR fragments were not amplifiable. Similar results were observed when purified DNA or amplified WGA product derived from these FFPE tissues were used directly in the multiplex qPCR assay (data not shown). (B) aCGH was performed to demonstrate a correlation between the multiplex PCR results and aCGH performance. 1 μg of WGA2 products were used for BAC aCGH analysis using PerkinElmer’s Spectral Labeling Kit and SpectralChip™ 2600 array platform per manufacturer’s recommendations. The ideograms below are representative of the data obtained with this sample set. They were generated using PerkinElmer’s Spectralware™ BAC array analysis software. High quality array statistics and QC metrics were obtained with samples 1 and 2, where as samples 3, 4 and 5 produced irregular array statistics and poor QC metrics. Test and control hybridization samples are labeled in Figure B.
Multiplex PCR Protocol for the Assessment of Damaged DNA (e.g. FFPE tissue DNA)
JumpStart™ REDTaq™ ReadyMix™ PCR reaction mix is recommended for this process. Reagents may be scaled proportionally if performing PCR reactions of smaller volume.
1. Prepare 10 μM MultiPlex Primer Mix by combining all ten primers in a suitable vessel at a final concentration of 10 μM each primer. Refer to Table 1 for primer sequences.
2. Combine the following reaction components in a suitable sized tube, per table 2 below. Scale-up master mix appropriately for the number of reactions being performed. Make extra master mix to account for pipetting loss. Note: The final PCR reaction volume may be scaled up or down as long as reagent concentrations are unchanged.
Multiplex PCR Master Mix |
|---|
3. Add 45 μL of the resulting master mix to an appropriate PCR tube or plate.
4. Add 5 μL of genomic DNA and mix until homogenous. For best results, use between 10 and 100 ng of template DNA per reaction. Note: Alternatively, 5 μL of undiluted GenomePlex WGA2 tissue lysate, or 100 ng of WGA amplicons generated with any of our GenomePlex products can be added instead of purified genomic DNA.
5. Place PCR tube(s) or plate in thermal cycler and amplify DNA using the following cycling parameters
- 94 °C for 2 minutes to denature
- 35 cycles of:
- 94 °C for 1 minute
- 60 °C for 1 minute
- 72 °C for 1 minute
- 72 °C for 7 minutes
- 4 °C hold
Resolve 5 μL of resulting amplicons on a 4% agarose gel. Note: JumpStart RedTaq ReadyMix contains gel-loading solution, which allows immediate sample loading onto an agarose gel after PCR. All five PCR amplicons (132 bp, 150 bp, 196 bp, 235 bp and 295 bp) will be generated with high quality genomic DNA. Low quality DNA may fail to produce amplicons, or may present as faint bands for some amplicons ( Figure 1).
Conclusions
In conclusion, the assay described here is useful in determining DNA quality from samples that potentially contain damage (e.g. FFPE tissues, serum/plasma, forensic samples, etc.). Although it is useful for determining DNA quality, it doesn’t afford quantitative information. In these cases, qPCR is a viable alternative to determine DNA concentration of damaged samples. 5,6 This assay cannot predict the usefulness of damaged DNA without prior validation work. Rather, one must perform correlative experiments to empirically determine a quality threshold, for their downstream application(s) of choice.
References
Um weiterzulesen, melden Sie sich bitte an oder erstellen ein Konto.
Sie haben kein Konto?