Strategies for Deglycosylating N-Linked Glycans
When researching with glycoproteins and glycopeptides, there are various strategies for deglycosylating N-linked glycans. These strategies can involve PNGase F, PNGase A, and even native and sequential deglycosylation with endoglycosidases and exoglycosidases. Read on to learn details about the different types of deglycosylation for N-linked glycans and find which strategy works best for your research.
PNGase F
The use of the endoglycosidic enzyme PNGase F (N-Glycosidase F) is the most effective method of removing nearly all N-linked oligosaccharides from glycoproteins. PNGase F cleaves all N-linked or asparagine-linked hybrid or complex and high mannose oligosaccharides unless the core contains an α(1→3)-fucose. The asparagine must be peptide bonded at both termini. A tripeptide with the oligosaccharide-linked asparagine as the central residue is the minimal substrate for PNGase F (Figure 1). Detergent and heat denaturation increases the rate of cleavage up to 100 times. Most native proteins can still be completely N-deglycosylated, but incubation time must be increased. The optimal pH is 8.6 and the enzyme is active in the pH range of 6 to 10.

Figure 1.Cleavage site and structural requirements for PNGase F.
R1 = N- and C-substitution by groups other than H
R2 = H or the rest of an oligosaccharide structure
R3 = H or α(1→6)fucose
The asparagine residue from which the glycan is removed is deaminated to aspartic acid (Figure 2). The oligosaccharide is left intact and is suitable for further analysis.

Figure 2.Cleavage products from PNGase F treatment of N-glycans.
Steric hindrance slows or inhibits the action of PNGase F on certain residues of glycoproteins. Denaturation of the glycoprotein by heating with SDS and 2-mercaptoethanol greatly increases the rate of deglycosylation. PNGase F is available as a solution and a lyophilized powder.
Proteomics Grade PNGase F
Proteomics Grade PNGase F is extensively purified and lyophilized from dilute potassium phosphate buffer to produce a stable product. The product is free from glycerol and other stabilizers that may interfere in sensitive glycoprotein analysis methods. Features include:
- Excellent for applications requiring N-linked deglycosylation (Figure 3).
- Superior performance for on-blot, in-gel, and in-solution digestion methods.
- High specific activity of ≥25,000 units/mg.
- Compatible for use in MALDI-TOF mass spectrometry.

Figure 3.SDS-PAGE analysis of native and PNGase F-treated α-1 antitrypsin. The test sample (Lane 5) was deglycosylated in solution with 5 units of PNGase F for 1 hour at 37 °C prior to separation on SDS-PAGE. Note the shift in the mobility of the band upon deglycosylation.
Lanes
1: Molecular weight marker
2, 3, 4: Control, native α-1 antitrypsin
5: In-solution deglycosylated α1 antitrypsin
This PNGase F is also available in the GlycoProfile™ I, In-Gel Deglycosylation Kit. This kit has been optimized to provide a convenient and reproducible method to N-deglycosylate and trypsin-digest protein samples from 1D or 2D polyacrylamide gel pieces for subsequent MS or HPLC analysis.
PNGase A (GLYCOPEPTIDASE A)
Oligosaccharides containing a fucose α(1→3)-linked to the asparagine-linked N-acetylglucosamine (GlcNAc), commonly found in plant glycoproteins or glycoproteins parasitic worms, are resistant to PNGase F. PNGase A, isolated from almond meal, must be used in this situation.
PNGase A (aka Glycopeptidase A or Peptide-N-Glycosidase A) hydrolyzes oligosaccharides containing a fucose residue α(1→3)-linked to the asparagine-linked N-acetylglucosamine (Figure 4). Like PNGase F, the asparagine residue from which the glycan is removed is deaminated to aspartic acid. However, PNGase A is ineffective when sialic acid is present on the N-linked oligosaccharide.
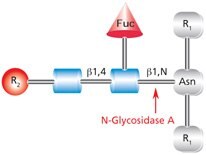
Figure 4.Cleavage site and structural requirements for PNGase A (Glycopeptidase A).
R1 = N- and C-substitution by groups other than H
R2 = H or the rest of an oligosaccharide structure
NATIVE AND SEQUENTIAL N-LINKED GLYCAN STRATEGIES
Glycoproteins may have buried or unavailable glycoconjugates. So, when removing glycans from a whole glycoprotein denaturation may be necessary to achieve full deglycosylation, usually with detergents that need to be sequestered before the enzymatic step. See example protocol by Freeze, H.1
If proteolytic digest is used, such as with a tryptic digest, the deglycosylation step will not require denaturation, but you may need to remove the protease to accommodate the deglycosylation enzyme.
Other commonly used endoglycosidases such as Endoglycosidase H and the Endoglycosidase F series are not suitable for general deglycosylation of N-linked sugars because of their limited specificities and because they leave one N-acetylglucosamine residue attached to the asparagine. These are useful for the deglycosylation of native proteins. Sequential deglycosylation can be done with exoglycosidases.
ENDOGLYCOSIDASE H
Endoglycosidase H (aka β-N-Acetylglucosaminidase H) cleaves the β-1,4 glycosidic linkage of high mannose N-linked glycans between the N-acetylglucosamine residues GlcNac1 and GlcNac2, also known as the chitobiose core (Figure 5). The cleavage leaves a glycoconjugate N-acetylglucosamine residue attached to the asparagine on the protein but removes the bulk of the glycan. This is useful especially for deglycosylation under native conditions, but the glycan may need to be processed with exoglycosidases to form high mannose glycoforms. Because the cleavage site is further removed from the amino acid sequence, it is thought to be more accessible to the enzyme.

Figure 5.Cleavage site and structural requirements for Endoglycosidase H (Endo H).
R1 = N- and C-substitution by groups other than H
R2 = H or the rest of an oligosaccharide structure
R3 = H or α(1→6)fucose
The specificity of this enzyme is such that oligomannose and most hybrid types of glycans, including those that have a fucose residue attached to the core structure, are cleaved whereas complex type glycans are not released. Thus, this enzyme is extremely useful for the selective release of high mannose (oligomannose) or hybrid type glycans from glycoproteins. The enzyme is also active against dolichol-linked glycans containing these structures.
The functional pH range for Endoglycosidase H is 5.0 to 6.0, with the optimum pH at 5.5. No loss of activity was observed during incubation at 37 °C for 48 hours over the pH range of 4.5 to 8.5. However, below pH 4.5, activity is rapidly lost. Endoglycosidase H is available as a native lyophilized powder, recombinant solution, or recombinant solution with reaction buffer.
ENDOGLYCOSIDASE F
Endoglycosidases F1, F2, and F3 are less sensitive to protein conformation than PNGase F and are more suitable for deglycosylation of native proteins. Endoglycosidase F1 (Endo F1) cleaves asparagine-linked or free high mannose (oligomannose) and hybrid structures, while Endoglycosidase F2 (Endo F2) and Endoglycosidase F3 (Endo F3) can cleave complex structures. The linkage specificities of Endoglycosidases F1, F2, and F3 suggest a general strategy for deglycosylation of proteins that may remove all classes of N-linked oligosaccharides without denaturing the protein. The Native Protein Deglycosylation Kit supplies all three of these enzymes (Endo F1, Endo F2, and Endo F3) with reaction buffers and detailed interactions.
Endoglycosidase F1
Endo F1 (aka Endo-β-N-acetylglucosaminidase F1) cleaves between the two N-acetylglucosamine residues in the N-linked diacetylchitobiose glycan core of the oligosaccharide, generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the asparagine (Figure 6).
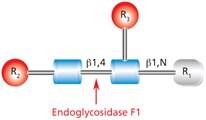
Figure 6.Cleavage site and structural requirements for Endoglycosidase F1.
R1 = H or Asn
R2 = Oligomannose or hybrid configuration
R3 = H or α(1→6)fucose
Endo F1 cleaves asparagine-linked or free high mannose (oligomannose) and hybrid structures, but not complex oligosaccharides (Figure 7). Core fucosylation of hybrid structures reduces the rate of cleavage by Endo F1 more than 50 fold. Endo F1 will cleave sulfated high mannose oligosaccharides whereas Endoglycosidase H will not. Endo F1 may be used under native or non-denaturing deglycosylation conditions.

Figure 7.Cleavage site for Endoglycosidase F1 in (A) high mannose glycans and (B) hybrid glycans.
Endoglycosidase F2
Endo F2 (aka Endo-β-N-acetylglucosaminidase F2) cleaves between the two N-acetylglucosamine residues in the N-linked diacetylchitobiose glycan core of the oligosaccharide, generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the asparagine (Figure 8).
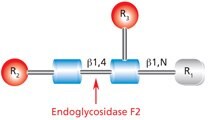
Figure 8.Cleavage site and structural requirements for Endoglycosidase F2.
R1 = H or Asn
R2 = Biantennary or oligomannose configurations
R3 = H or α(1→6)fucose
Endo F2 cleaves biantennary complex oligosaccharides (Figure 9). High mannose oligosaccharides are cleaved at a rate reduced 20-fold compared to that of complex structures. Endo F2 will not cleave hybrid structures. Fucosylation has little effect on Endo F2 cleavage of biantennary structures. Endo F2 is useful under native or non-denaturing deglycosylation conditions.
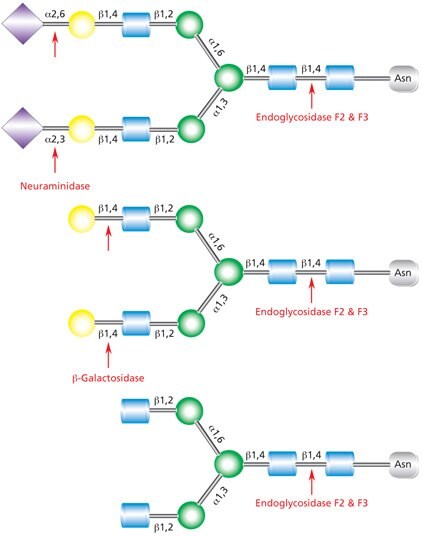
Figure 9.Cleavage site for Endoglycosidases F2 and F3 in a complex biantennary glycan. Sequential degradation using the exoglycosidic enzymes neuraminidase (top) and β-galactosidase (middle) to remove terminal monosaccharides is shown.
Endoglycosidase F3
Endo F3 (aka Endo-β-N-acetylglucosaminidase F3) cleaves between the two N-acetylglucosamine residues in the N-linked diacetylchitobiose glycan core of the oligosaccharide, generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the asparagine (Figure 10).
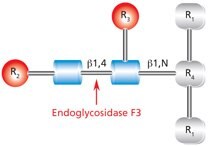
Figure 10.Cleavage site and structural requirements for Endoglycosidase F3.
R1 = N- and C-substitution by groups other than H
R2 = Biantennary or triantennary complex oligosaccharide or trimannosyl (Man3) core
R3 = H or α(1→6)fucose
R4 = Asn (Asn or H if fucosylated at R3)
Endo F3 has no activity on oligomannose and hybrid molecules. Endo F3 cleaves non-fucosylated biantennary and triantennary complex oligosaccharides at a slow rate, but only if peptide-linked (Figures 9 and 11). Core fucosylation of biantennary structures increases activity up to 400 fold. Core fucosylated biantennary structures are efficient substrates for Endo F3, even as free oligosaccharides. Endo F3 will also cleave fucosylated trimannosyl core structures on free and protein-linked oligosaccharides.

Figure 11.Cleavage site for Endoglycosidases F3 in complex triantennary glycan. Sequential degradation using the exoglycosidic enzymes neuraminidase (top) and β-galactosidase (middle) to remove terminal monosaccharides is shown.
Native deglycosylation of complex tetraantennary glycans requires sequential hydrolysis down to the trimannosyldiacetylchitobiose (Man3GlcNAc2) core prior to cleavage using Endo F3 (Figure 12).
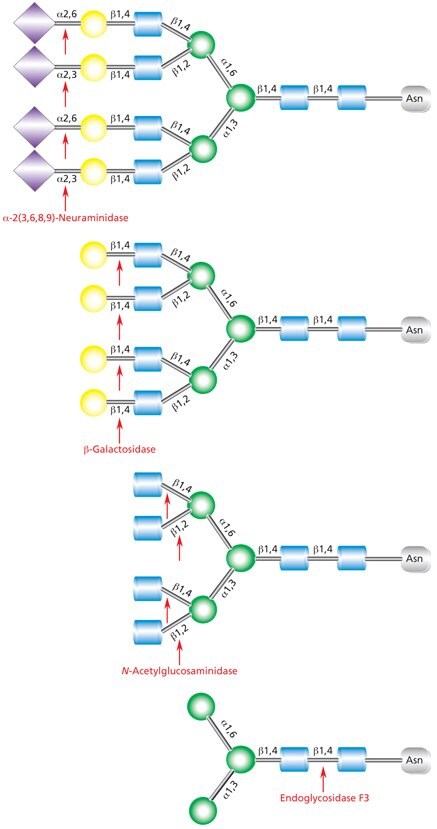
Figure 12.Sequential deglycosylation of complex tetraantennary glycan. The terminal monosaccharides are sequentially removed using the exoglycosidic enzymes neuraminidase (top), β- galactosidase (middle), and N-acetylglucosaminidase (bottom), until the trimannosyldiacetylchitobiose core remains for subsequent cleavage by Endo F3.
EXOGLYCOSIDASES
Through sequential deglycosylation of monosaccharides using exoglycosidases, all complex oligosaccharides can be reduced to the trimannosyldiacetylchitobiose (Man3GlcNAc2) core. Complex N-linked glycans can be selectively hydrolyzed with a neuraminidase, β-galactosidase, and N-acetylglucosaminidase, available as part of the Enzymatic Deglycosylation Kit. Additional cleavage using fucosidases may be required in some situations. The remaining trimannosyldiacetylchitobiose core structures can be removed with Endoglycosidase F3.
References
Zaloguj się lub utwórz konto, aby kontynuować.
Nie masz konta użytkownika?