esiRNA FAQs (Frequently Asked Questions)
MISSION® esiRNA are endoribonuclease prepared siRNA that consists of a heterogeneous mixture of siRNA that all target the same mRNA sequence. These multiple silencing triggers lead to highly specific and effective gene silencing. Here are some of the most commonly asked questions regarding esiRNA uses and availability.
How many screenings can be performed with the 1, 2.5, and 5 µg quantities of esiLibrary and esiFLEX?
Depending on the cell line, 30 to 200 ng or 9 to 60 ng of esiRNA are needed per 96-well and 384-well plate well, respectively. Assuming a 90% success rate, the following number of transfections are probable:
What if my favorite gene is not on the list of Individual esiRNA?
We are constantly adding new Individual esiRNA based on updated genome annotations and expression validation. For transcripts that are not covered, we offer custom-made esiRNA (this product option is called, esiOPEN) that are fully flexible in terms of species and target sequence. A target with a minimum length of 500 bp is required for the esiOPEN design process.
Are esiRNA available for species other than human or mouse?
Yes, we offer custom-made esiRNA (this product option is called, esiOPEN) that are independent of the origin of the sequences. You only must provide a target sequence with a minimum length of 500 bp.
Does esiRNA target all transcript variants of the target-gene?
Yes, esiRNA are designed to a region of the target that is common to all transcript variants. However, in rare cases a variant is significantly different to all others so that no common region can be found. In these cases, two esiRNA are offered: one that targets the transcripts that share a common region and another that targets the version not covered with the first esiRNA.
Are esiRNA available that target individual transcript variants of the same gene independently?
Yes, we offer custom-made esiRNA (this product option is called, esiOPEN) for any target-region. A target with a minimum length of 500 bp is required for the esiOPEN design process. Therefore, the target variant must be different from the other variants by at least these 500 bp to be specific.
How can I validate an observed RNAi phenotype?
RNAi phenotypes can be validated utilizing independent, secondary esiRNA that target a region of the transcript different from the primary esiRNA. The esiRNA for this purpose are available as the product option called, esiSEC.
What can be done if the knockdown is not strong enough?
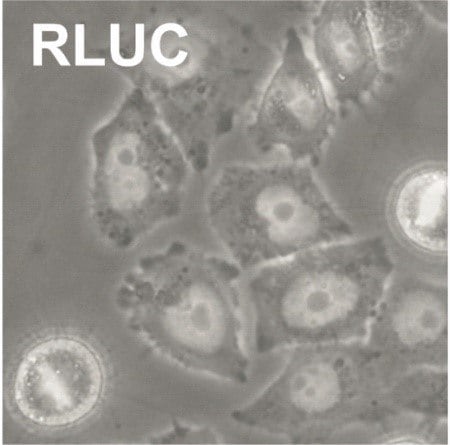
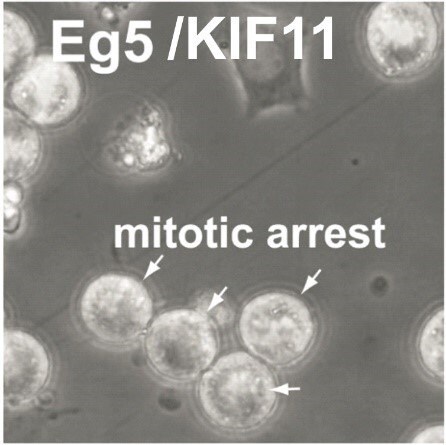
A possible reason for a weak knockdown is low knockdown efficiency. Optimization of the reaction should be performed with a dose-response curve for the transfection reagent and esiRNA. The amount of transfection reagent should be determined using a negative control such as Renilla Luciferase (RLUC). For validation of knockdown efficiencies use a positive control, e.g. Eg5, and perform phenotypic analysis of mitotic arrest (rounded cells) or qPCR analysis. Cell numbers must be kept stable for all transfections as differences from the optimized condition will affect knockdown efficiencies or cause cytotoxic effects.
If transfection was optimized and of good quality, many other reasons can lead to an unsatisfactory depletion of the target protein. For proteins that have a slow turn-over rate, the maximum depletion can be expected 96 hours post-transfection, e.g. the APC subunit of Cdc27. If the cell line divides slowly, this may also lead to longer knockdown delays. If the protein turn-over rate is high, the depletion can be expected 48 hours post-transfection, e.g. the motor protein Eg5. Therefore, measurements must be made at the right time point. However, in some cases, if poor knockdown efficiency cannot be explained, it might worthwhile to try an independent, secondary esiRNA (this product option is called, esiSEC) to target another region in the mRNA.
Can I use the same protocol for the transfection of siRNA and esiRNA?
No, while the transfection for siRNA and esiRNA are comparable, they are not identical. An siRNA transfection protocol may serve as a starting point for the separate esiRNA optimization process. Cell lines have different properties and are sensitive or insensitive to different manipulations. This is certainly also true for transfections. Therefore, for every cell line, an appropriate transfection protocol must be established separately. The goal of transfection optimization is to achieve maximal transfection efficiency with minimal cytotoxicity.
Provided below are the esiRNA transfection conditions for some commonly used cell lines. The values provided should only serve as guidelines (a detailed optimization should be carried out if other plate formats are used). We recommend using an esiRNA for Eg5 (KIF11) and Renilla luciferase (RLUC) as positive and negative controls, respectively, for optimization. Eg5 (KIF11) induces mitotic arrest with the formation of round-shaped cells that become apoptotic after several hours of arrest. These round-shaped cells can easily be observed in a standard bright-field cell-culture microscope. The conditions with a maximum number of round-shaped cells for the Eg5 transfection with a minimum cytotoxicity for the RLUC transfection signify optimal transfection conditions.
Zaloguj się lub utwórz konto, aby kontynuować.
Nie masz konta użytkownika?