Workflow for High-Throughput Glycoprofiling of Rituximab
Stephan Altmaier1, Pegah Jalili2, Uma Sreenivasan3, Kevin Ray2
1Darmstadt, Germany, 2St. Louis, MO, USA, 3Round Rock, TX, USA
Protocol for purification, reduction, and SEC-MS glycoform analysis of a therapeutic monoclonal antibody

Antibody Purification
Antibody Reduction
Chromatography
Measurement & Analsysis
Abstract
A complete SEC-MS workflow has been developed to enable rapid glycoprofiling of monoclonal antibodies in cell culture supernatants. In detail, it includes:
- Antibody purification using immobilized protein A
- Antibody reduction procedure
- Mass spectrometer calibration
- System suitability test utilizing a recombinant human monoclonal antibody reference
- SEC-MS method for sample separation and analysis
Introduction to the Chracterization of Antibody-based Drugs
Monoclonal antibodies (or immunoglobulins - IgGs) are large glycoproteins with a molecular weight of approximately 150 kDa (150,000 g/mol). They are composed of two light chains (LC, molecular weight ca. 25 kDa each) and two heavy chains (HC, molecular weight ca. 50 kDa each) linked through covalent inter-chain disulfide bonds. They are utilized for the treatment of various types of cancer, and other diseases such as multiple sclerosis, Alzheimer’s disease, and migraine.
Careful and thorough characterization of therapeutic mAbs is essential for ensuring drug safety and efficacy. MAbs are typically manufactured in mammalian host cell lines in bioreactors, generating a large number of heterogeneous drug molecules. Establishing a number of critical quality attributes (CQAs) for each mAb and demonstrating that production batches are within acceptable limits are requirements for both innovator and biosimilar therapeutics.1,2
In many cases, the characterization of an antibody-based drug is performed using a specific chromatographic technique (e.g., size exclusion, reversed phase or hydrophilic interaction liquid
chromatography, respectively – SEC, RP or HILIC)3,4 coupled with mass spectrometry (MS). This combination allows for different types of analyses to be carried out, e.g., accurate mass measurement of the intact mAb and subunits, peptide mapping, and the determination of post-translation modifications such as glycosylation, oxidation, and deamidation.
Several techniques are applied to simplify antibody analysis by either fragmentation or removal of glycans. The latter can be performed by treatment with PNGase F, whereas proteolysis with IdeS5 or reduction of inter-chain disulfide bonds with reducing agents, such as dithiothreitol, result in the formation of different antibody fragments with masses of 25– 50 kDa. Various combinations of these techniques can be applied. For analysis of mAbs in cell culture supernatants, these may be combined with a preceding affinity purification step.6 These approaches are referred to as intact mass and middle-up analysis methods.7 The former term relates to the measurement of the mass of an intact mAb without controlled dissociation being performed. Such an experiment reveals information about stoichiometry, proteoforms, and modifications. Middle-up experiments include mass measurement after cleaving mAbs into several large fragments/ subunits via chemical reduction or proteolytic digestion. An example of this approach is the analysis of mAb light and heavy chains, providing insight into post- translational modifications of the individual chains. Figure 1 provides an overview of antibody sample preparation and various digestion options prior to middle-up mass analysis.
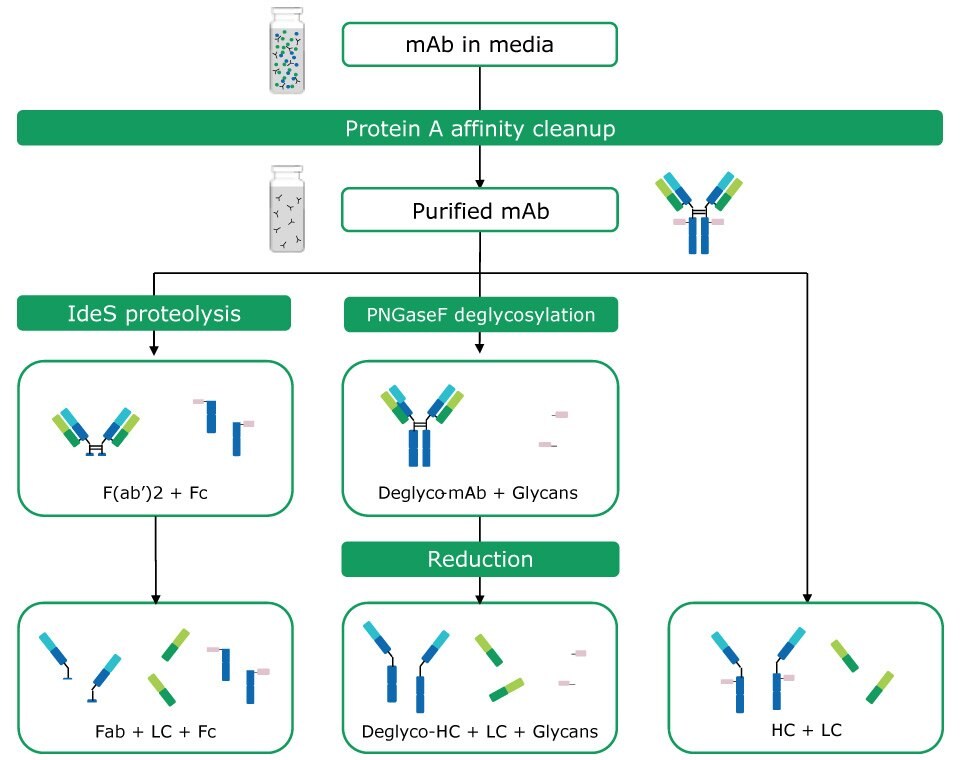
Figure 1.Antibody sample preparation by protein A affinity cleanup and chemical and proteolytic digestion options: Proteolysis with IdeS (formation of Fc and Fab fragments); PNGase F treatment (deglycosylation); chemical reduction (release of heavy and light chains). A combination of proteolysis and chemical reduction is also possible.
Glycosylation is one of the most common and important post translational modifications for mAbs. Glycans attached to antibodies play an important role in their pharmacokinetics, efficacy, and safety. Glycosylation involves the attachment of glycans at specific sites on a protein, most commonly at asparagine (Asn, N-linked) or serine/threonine (Ser/Thr, O-linked) amino acid residues. Both types of glycosylation are important for protein conformation, protein activity, protection from proteolytic degradation, and intracellular trafficking and secretion. Based on the large influence of glycosylation on protein function an accurate study and analysis of glycans is essential. N-glycan composition can be determined by the analysis of four different target structures: intact glycoproteins, glycopeptides, released glycans, and monosaccharide analysis.
This report describes the generation of heavy chain mass data for the rapid glycoprofiling of reduced rituximab recombinant monoclonal antibody samples expressed in 30 separate CHO cell clones along with an authentic rituximab reference material. Samples were purified using protein A resin, reduced, and analyzed by SEC-MS. Deconvoluted heavy chain spectra were generated, and glycoform relative distributions were determined. In all experiments, the recombinant human monoclonal antibody SILu™Lite SigmaMAb™ (#MSQC4) was utilized as a reference antibody.
Procedure for mAb Purification, Reduction, and SEC-MS Glycoform Analysis
The target antibody purification was performed on cell culture supernatants using immobilized protein A resin in a 96-well format. The suggested minimum working mAb titer is 100 µg/mL. All procedures were conducted using both reference and assay control samples of SigmaMAb™ with a molecular mass of ~150 kDa. The reference sample consisted of the pure antibody reconstituted in water; it is used for the system suitability tests and instrument check. The assay control sample contained the antibody spiked into or delivered as a mixture with cell culture media or spent media (cell broth including nutrients etc.); it goes through the entire workflow and functions as a control sample.
mAb Sample Prepartion and System Setup
In detail, the high-throughput purification of mAbs from cell culture media using protein A resin was performed as follows:
System/Workflow suitability
As part of the workflow suitability, an assay control of SigmaMAb™ in media was purified along with the samples. Reference sample (SigmaMAb™) is prepared as follows:
- Reconstitute each vial of MSQC4 in 1.0 mL water to obtain a solution with an antibody concentration of 1 mg/mL.
- Prepare assay control (spent media sample) by spiking SigmaMAb™ in EX-CELL® CHOZN®
- Platform Medium, or equivalent, to obtain a final concentration of 100-500 µg/mL.
Preparation of equilibration and elution buffers
- Prepare equilibration buffer (20 mM citrate, 150 mM NaCl, pH 7) by dissolving 5.82 g trisodium citrate dihydrate, 0.04 g citric acid, and 8.77 g sodium chloride in 1 L water. Adjust pH of resulting solution to 7 using 1 M NaOH or HCl as needed; subsequently filter solution using a 0.2 µm filter.
- Prepare elution buffer (25 mM citrate, pH 3) by dissolving 4.8 g citric acid in 1 L water. Adjust pH of resulting solution to 3 using 1 M NaOH or HCl as needed.
Clarify samples
- Centrifuge samples in tubes at maximum speed for five minutes and samples in plates at maximum speed for 60 minutes.
Protein A loading
- Add or remove water from top portion of settled protein A slurry to obtain a 50% protein A suspension.
- Mix slurry by constant pipette action and gentle shaking of reagent reservoir.
- Use a multichannel pipette to deliver 200 µL of protein A slurry to each well of a 96-well filter plate. Place protein A filter plate on a vacuum manifold. Catch any flow-through from the filter plate by placing the filter plate on top of a used collection plate.
Protein A equilibration
- Add 200 µL of equilibration buffer to each well of protein A and apply vacuum to void wells of buffer.
- Repeat both steps twice.
mAb binding
- Remove 750 μL of solution of sample and control, without disturbing the pellet, and load plate.
- Cover the plate with film and secure filter and collection plates with a rubber band.
- Incubate on an orbital shaker at 170 rpm for 30 minutes.
Washing bound mAb
- Place protein A filter plate on vacuum manifold with a waste collection plate inserted and the film cover removed.
- Apply vacuum to void wells of buffer media and transfer the filter plate onto a waste collection plate.
- Add 200 µL of equilibration buffer to wells and centrifuge plates at 3700 rpm for five minutes (this step helps in clearing the sample film on sides of filter plate wells).
- Add 200 µL of equilibration buffer to wells and apply vacuum to remove buffer.
- Repeat once more for a total of three washes.
Eluting bound mAb
- Place protein A filter plate on a new collection plate and secure with a rubber band.
- Add 100 µL of elution buffer to each well, incubate filter plate on orbital shaker at 170 rpm for five minutes.
- Centrifuge plates at 3700 rpm for five minutes.
- Repeat addition of elution buffer, incubation on orbital shaker, and centrifugation for a total of three elution steps (300 µL of total elution volume).
Typical antibody recovery using this procedure is 60%.
Antibody Reduction Procedure
Partial disulfide (S-S) bond reduction was performed as follows:
- Prepare a 1 M dithiothreitol (DTT) solution by dissolving 154.25 mg DTT in 1 mL water.
- Prepare a 1 M ammonium bicarbonate (ABC) solution by dissolving 79.06 mg ABC in 1 mL water.
- Combine equal volumes of 1 M ABC and 1 M DTT to prepare the reduction solution.
- Transfer aliquots of 50 μL of each sample, system suitability reference, and control to autosampler vials.
- Reduce by addition of 5 μL 0.5 M ABC/0.5 M DTT solution.
- Incubate for one hour at room temperature or 30 min at 37 °C.
Note: Partial reduction is performed under non- denaturing conditions, where the inter-chain disulfide bonds (which are more susceptible to reduction) will break and produce the light and heavy chains, while the intra-chain disulfide bonds within each individual domain remain intact.
Alternatively, complete reduction of samples can be performed by using this protocol:
- Prepare a 100 mM solution of tris(2-carboxyethyl) phosphine (TCEP) in 6 M aqueous guanidine hydrochloride by dissolving 2.87 g of TCEP and 57.32 g of guanidine hydrochloride in 100 mL of water (if less solution is needed, scale down accordingly).
- Combine 30 μL of the resulting solution with 10 μL of sample.
- Incubate for two hours at 37 °C.
Note: Complete reduction is performed under denaturing conditions, where both the inter-chain and intra-chain disulfide bonds will break.
Instrumentation Calibration for mAb Analysis
The Waters™ QToF Xevo® G2XS mass spectrometer was calibrated in a mass range of 500 – 6000 m/z with a 20 μL/min infusion of 0.4 mg/mL of cesium iodide in water. Alternatively, calibration can be performed with a 20 μL/min infusion of 0.4 mg/mL of polyalanine in water prior to running the samples.
System Suitability for mAb Analysis
To evaluate performance of the entire workflow, an assay control (SigmaMAb™ in media) was prepared and analyzed along with the samples. Reduced SigmaMAb™ reference was also tested to ensure system suitability (see section above).
SEC-MS System Setup and MS Data Analysis for mAb Glycoforms
1. SEC-MS system setup
The essential settings of the UHPLC-PDA chromatography system and the qToF mass spectrometer applied in the analysis of reduced antibodies are listed in Table 1 and Table 2 below.
2. MS mAb Glycoform Data Analysis
Data were processed using the MaxEnt1 module within the MassLynx™ 4.1 software to generate and analyze deconvoluted (zero charged) mass spectra.
In general, a summed spectrum was created from the corresponding total ion chromatogram (TIC) of the eluted heavy chain (HC). The summed m/z spectrum was then processed by the MaxEnt1 algorithm; detailed parameters are listed in Table 3.
For glycoform analysis, data were processed using UNIFI software from Waters™. Glycoforms were matched by the software and HC glycoform glycans are listed in the respective section below. Glycoform relative abundance data were tabulated based on peak intensities of the co-eluting glycoform species. A deconvolution filter setting employing a base peak intensity of 2% was utilized to preclude noise incorporation, and an output resolution setting of 5 Da was used
Results of mAb purification, reduction, and SEC-MS glycoform analysis
The analysis objective was to perform rapid glycoprofiling of a set of reduced rituximab samples by SEC-MS. 30 cell culture supernatant samples from separate CHO clones expressing rituximab were received in 1.5 mL microcentrifuges tubes and stored at -20 °C prior to analysis. An authentic rituximab sample was also tested for comparison. In addition, SigmaMAb™ antibody reference and control samples were used to determine system suitability.
Protein A purification and reduction of rituximab samples and SigmaMAb™ antibody control was performed as described in the previous section. Purification was performed on 500 μL of each sample and 750 μL of SigmaMAb™ control. SigmaMAb™ reference underwent reduction only (no protein A purification) and was used to determine system suitability.
Subsequently, samples were analyzed in their reduced form via SEC-MS.
Table 4 and Table 5 list all glycans searched in this work and glycan constituent monosaccharides, respectively.
System suitability test results
10 μL of a reduced SigmaMAb™ reference sample was injected on the SEC-PDA-MS system. Figure 2 displays the photodiode array (280 nm) and TIC traces of the SigmaMAb™ reference, while Figure 3 shows the summed and deconvoluted mass spectra of the reduced SigmaMAb™ reference heavy chain glycoforms. Obtained data demonstrated that the system and assay were suitable for glycoform mass analysis. The system reliably measured glycoform distributions throughout the acquisition queue, with control data falling within historical ranges for relative composition of G0F and G1F glycoforms and indicating proper calibration and sample reduction conditions. In more detail, the observed heavy chain mAb glycoform masses matched the expected masses of SigmaMAb™, as listed in Table 6. Discrepancies between the observed masses and the theoretical values for three glycoforms are all within 0.003% mass error or less.
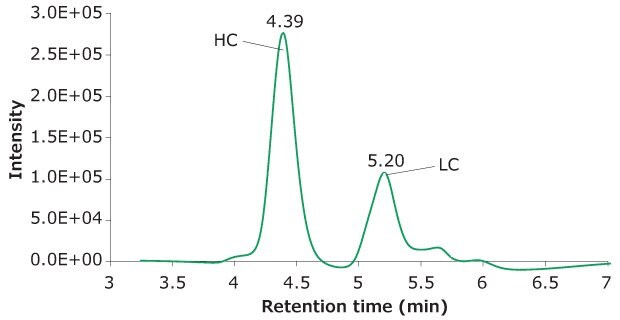
Figure 2A.Photodiode array (280 nm).
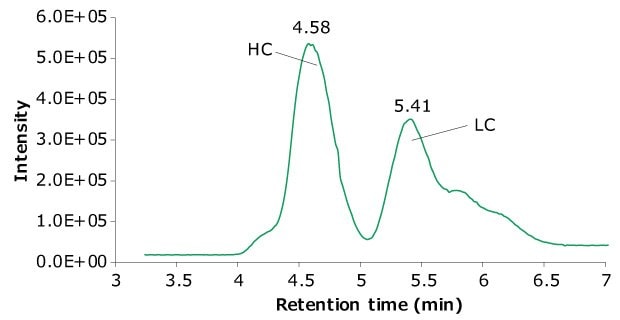
Figure 2B.TIC trace of reduced SigmaMAb™ reference.
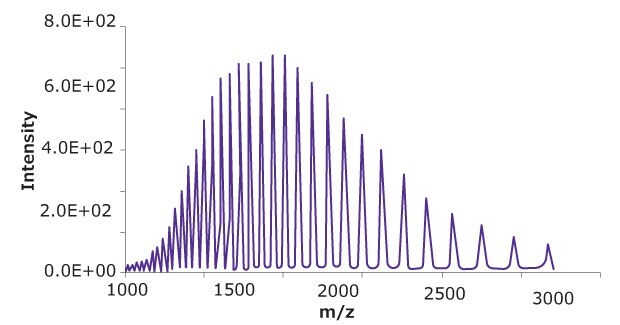
Figure 3A.Summed mass spectra of heavy chain of reduced SigmaMAb™ reference.
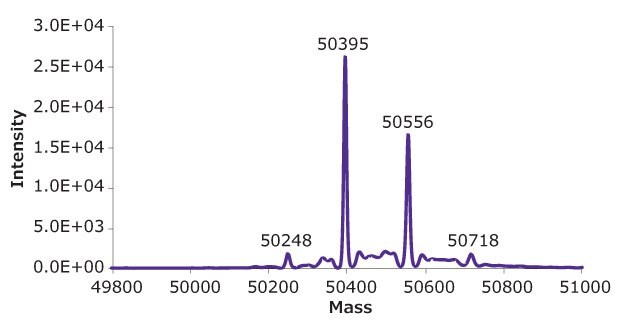
Figure 3B.Deconvoluted mass spectra of heavy chain of reduced SigmaMAb™ reference.
Rituximab monoclonal antibody sample test results
30 rituximab monoclonal antibody samples from separate CHO clones were analyzed for their glycoform composition by SEC-MS along with an authentic rituximab reference. Figure 4 displays deconvoluted spectra of the glycoform composition of two exemplary antibody samples (clones 8 and 27). Typical protein adducts surrounding the major species are at low but appreciable levels; hence, care should be taken when considering glycoforms with composition values less than 3%. Table 7 and Table 8 show observed and theoretical masses of the two reduced rituximab samples from clones 8 and 27. The mass errors between the observed masses and the theoretical values for the different glycoforms are in the range of 0.004 to 0.023%.

Figure 4A.Exemplary deconvoluted SEC-MS spectra of rituximab from clones 8. The MS peak annotations correspond to glycoform glycans listed in Table 7 and Table 8.

Figure 4B.Exemplary deconvoluted SEC-MS spectra of rituximab from clones 27. The MS peak annotations correspond to glycoform glycans listed in Table 7 and Table 8.
Table 8 summarizes the results for all 30 samples and authentic reference and Table 9 provides an overview over the glycan composition of all 30 clones analyzed. G0F, G1F and G2F were the main glycans observed. In addition, a small fraction of the samples was also bearing G0, G0F-N and Man glycans.
Conclusion
The objective of this work was to develop a CHO clone screening workflow using an SEC-MS middle- up mass analysis of reduced rituximab recombinant monoclonal antibody samples, to generate protein mass data for the antibody heavy chains and to determine their glycoform compositions. Samples were purified from spent media, reduced, analyzed by SEC-MS and deconvoluted heavy chain spectra were generated for the determination of glycoform relative distributions.
The workflow comprises of an antibody purification process using immobilized protein A, a mAb reduction procedure, a mass spectrometer calibration method and a system suitability test utilizing SILu™Lite SigmaMAb™ Universal Antibody Standard human. In addition, an SEC method suitable for sample separation and analysis of reduced mAbs was utilized. A total of 30 rituximab cell culture supernatants from separate CHO clones underwent a protein A purification and a reduction procedure and were then subjected to SEC-MS analysis, along with an authentic rituximab reference. In addition, SigmaMAb™ was utilized as a reference and was treated in the same way.
Experiments utilizing a reduced SigmaMAb™ reference sample revealed that the observed mAb heavy chain glycoform masses matched the expected masses of the antibody fragments. In this case, discrepancies between the observed and the theoretical values for three glycoforms are all within 0.003% mass error or less, proving the system and assay were suitable for middle-up mass analysis. Control data falling within historical ranges for relative composition of G0F and G1F glycoforms indicated proper calibration and sample reduction conditions. Glycoform composition of rituximab from 30 CHO clones was determined by SEC-MS. Results reveal significant variation in the glycoprofiles of individual
clones, with G0F being the most abundant glycan for all clones screened in this experiment. The most abundant glycan observed in the authentic reference rituximab was G1F.
All generated data display that the implemented workflow can be used for reduced monoclonal antibody sample glycoform analysis, with accurate results allowing for an unambiguous identification and relative quantitation of six different glycans.
References
Zaloguj się lub utwórz konto, aby kontynuować.
Nie masz konta użytkownika?