추천 제품
포장
kit of 6 vials (reagents for 25 Reactions)
배송 상태
dry ice
저장 온도
−20°C
일반 설명
The T7 Endonuclease Detection Assay is a well-known method for detecting genome editing events from CRISPR, Zinc-finger nuclease, and TALEN gene targeting. Originally identified from Escherichia coli bacteriophage, the T7 endonuclease can cleave mismatched heteroduplex DNA, Holliday junctions, branched DNA, and cruciform DNA.
Following a gene editing experiment, genomic DNA surrounding the target locus is amplified by PCR, and the PCR amplicons are denatured and reannealed through heating and slow cooling. If NHEJ events have occurred, then, after reannealing, several products are possible. Homoduplexes can form where a WT strand is reannealead to a WT strand or an indel-carrying strand is reannealed to an indel-carrying strand. Heteroduplexes form when a WT strand is reannealed to an indel-carrying strand causing a mismatch. Heteroduplex products with mismatches are cleaved by the T7 endonuclease. Separating the DNA products after treatment with T7 endonuclease by gel electrophoresis will result in a banding pattern indicative of the amount of heteroduplexes in the sample. The amount of cleaved heteroduplexes is directly related to the amount of indel activity.
Following a gene editing experiment, genomic DNA surrounding the target locus is amplified by PCR, and the PCR amplicons are denatured and reannealed through heating and slow cooling. If NHEJ events have occurred, then, after reannealing, several products are possible. Homoduplexes can form where a WT strand is reannealead to a WT strand or an indel-carrying strand is reannealed to an indel-carrying strand. Heteroduplexes form when a WT strand is reannealed to an indel-carrying strand causing a mismatch. Heteroduplex products with mismatches are cleaved by the T7 endonuclease. Separating the DNA products after treatment with T7 endonuclease by gel electrophoresis will result in a banding pattern indicative of the amount of heteroduplexes in the sample. The amount of cleaved heteroduplexes is directly related to the amount of indel activity.
애플리케이션
Functional Genomics; Target Validation; Genome Editing
특징 및 장점
- Technically simple method based on well-known techniques
- Easily interpretable results
- Fast analysis turnaround
- Cost-effective
성분
Each kit consists of:
- one vial of T7 Endonuclease I
- one vial of Control Template and Primer Mix
- one vial of Buffer solution
- one vial of DNA Ladder - 1KB
- one vial of Gel Loading Dye (6X)
- one vial of Proteinase K
원리
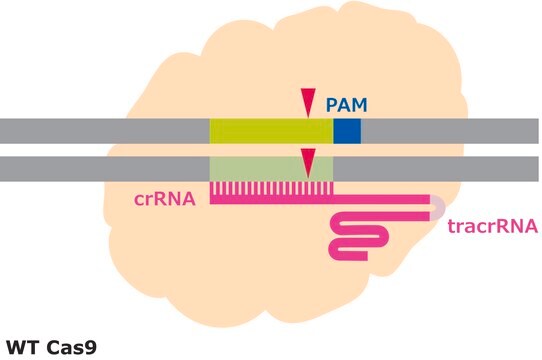
CRISPR/Cas systems are employed by bacteria and archaea as a defense against invading viruses and plasmids. Recently, the type II CRISPR/Cas system from the bacterium Streptococcus pyogenes has been engineered to function in eukaryotic systems using two molecular components: a single Cas9 protein and a non-coding guide RNA (gRNA). The Cas9 endonuclease can be programmed with a gRNA, directing a DNA double-strand break (DSB) at a desired genomic location. Similar to DSBs are also induced by Zinc-finger nucleases (ZFNs) and TALENs. The cell then activates endogenous DNA repair processes, either non-homologous end joining (NHEJ) or homology-directed repair (HDR), to heal the targeted DSB.
Efficiency in gene editing can vary in large part due to the target sequences. Chromatin structure and some sequence elements, for example high GC-content, can inhibit editing at some genomic sequences, affecting sgRNA activity. Additionally, favorable bases in the sgRNA sequence such as a guanine proximal to the PAM can promote sgRNA activity, but these preferred bases may not be available at the target site. It is important to evaluate the gene editing ability of several sgRNAs by quantifying the frequency of modifications using a method like T7 endonuclease mismatch detection.
Efficiency in gene editing can vary in large part due to the target sequences. Chromatin structure and some sequence elements, for example high GC-content, can inhibit editing at some genomic sequences, affecting sgRNA activity. Additionally, favorable bases in the sgRNA sequence such as a guanine proximal to the PAM can promote sgRNA activity, but these preferred bases may not be available at the target site. It is important to evaluate the gene editing ability of several sgRNAs by quantifying the frequency of modifications using a method like T7 endonuclease mismatch detection.
신호어
Danger
유해 및 위험 성명서
예방조치 성명서
Hazard Classifications
Resp. Sens. 1
Storage Class Code
12 - Non Combustible Liquids
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
관련 콘텐츠
The largest offering of quality Cas9 proteins ensures you have the perfect CRISPR reagents for your specific experimental needs. Rigorous purification, NLS signals, increased specificity and activity ensure your gene editing experiments are efficient and simple.
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.