추천 제품
양식
solution
포장
pkg of 20,000 U (10703753001 [40 U/μl])
pkg of 5,000 U (10899208001 [10 U/μl])
제조업체/상표
Roche
파라미터
30 °C optimum reaction temp.
배송 상태
dry ice
저장 온도
−20°C
일반 설명
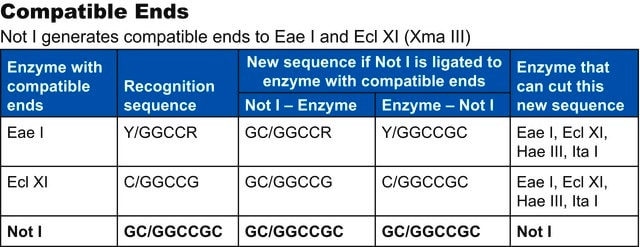
Compatible ends
Apa I ends are not compatible with those generated by any other known restriction enzymes.
Isoschizomers
Apa I is an isoschizomer to Bsp 120 I and Psp OMI.
Methylation sensitivity
Apa I is inhibited by 5′-methylcytosine at the sites indicated (*) in the recognition sequence.
Activity in SuRE/Cut Buffer System
Buffer printed in bold face type is the buffer recommended for optimal activity:
ABLMH
100%10-25%50-75%50-75%0-10%
Relative activity in complete PCR mix
Relative activity in PCR mix (Taq DNA Polymerase buffer) is 100%. The PCR mix contained target DNA, primers, 10 mM Tris-HCl (pH 8.3, +20°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 2.5 U Taq DNA polymerase. The mix was subjected to 25 amplification cycles. Activity in reaction buffer of Pwo SuperYield DNA Polymerase PCR Mix is 10%. When supplemented with GC-RICH Solution activity is increased to >100%.
Incubation temperature
+30°C
Unit definition
One unit is the enzyme activity that completely cleaves 1 μg ? × Hind III fragments in 1 hour at +30°C in a total volume of 25 μl SuRE/Cut Buffer A.
Heat inactivation
The enzyme can be heat inactivated by incubating it for 15 minutes at +65°C.
Number of cleavage sites on different DNAs
λAd2SV40φ X174M13mp7M13mp18pBR322pBR328pUC18
112 1000000
Ligation and recutting assay
Apa I fragments obtained by complete digestion of 1 μg λ × Hind III fragments are ligated with 1 U T4 DNA Ligase in a volume of 10 μl by incubation for 16 hours at +4°C in 66 mM Tris-HCl, 5 mM MgCl2, 5 mM Dithiothreitol, 1 mM ATP, pH 7.5 (at +20°C) resulting in >95% recovery of 1 μg λ × Hind III fragments.
Subsequent re-cutting with Apa I yields >95% of the typical pattern of λ × Hind III× Apa I fragments.
Apa I ends are not compatible with those generated by any other known restriction enzymes.
Isoschizomers
Apa I is an isoschizomer to Bsp 120 I and Psp OMI.
Methylation sensitivity
Apa I is inhibited by 5′-methylcytosine at the sites indicated (*) in the recognition sequence.
Activity in SuRE/Cut Buffer System
Buffer printed in bold face type is the buffer recommended for optimal activity:
ABLMH
100%10-25%50-75%50-75%0-10%
Relative activity in complete PCR mix
Relative activity in PCR mix (Taq DNA Polymerase buffer) is 100%. The PCR mix contained target DNA, primers, 10 mM Tris-HCl (pH 8.3, +20°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 2.5 U Taq DNA polymerase. The mix was subjected to 25 amplification cycles. Activity in reaction buffer of Pwo SuperYield DNA Polymerase PCR Mix is 10%. When supplemented with GC-RICH Solution activity is increased to >100%.
Incubation temperature
+30°C
Unit definition
One unit is the enzyme activity that completely cleaves 1 μg ? × Hind III fragments in 1 hour at +30°C in a total volume of 25 μl SuRE/Cut Buffer A.
Heat inactivation
The enzyme can be heat inactivated by incubating it for 15 minutes at +65°C.
Number of cleavage sites on different DNAs
λAd2SV40φ X174M13mp7M13mp18pBR322pBR328pUC18
112 1000000
Ligation and recutting assay
Apa I fragments obtained by complete digestion of 1 μg λ × Hind III fragments are ligated with 1 U T4 DNA Ligase in a volume of 10 μl by incubation for 16 hours at +4°C in 66 mM Tris-HCl, 5 mM MgCl2, 5 mM Dithiothreitol, 1 mM ATP, pH 7.5 (at +20°C) resulting in >95% recovery of 1 μg λ × Hind III fragments.
Subsequent re-cutting with Apa I yields >95% of the typical pattern of λ × Hind III× Apa I fragments.
Quality
Absence of nonspecific endonucleases
1 μg λ × Hind III fragments is incubated for 16 hours in 50 μl SuRE/Cut Buffer A with excess Apa I. The number of enzyme units which do not change the enzyme-specific pattern is stated in the certificate of analysis.
Absence of exonuclease activity
Approximately 5 μg [3H] labeled calf thymus DNA are incubated with 3 μl Apa I for 4 hours at +37°C in a total volume of 100 μl 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithioerythritol, pH approximately 7.5. Under these conditions, no release of radioactivity is detectable, as stated in the certificate of analysis.
Absence of nonspecific endonucleases
1 μg λ × Hind III fragments is incubated for 16 hours in 50 μl SuRE/Cut Buffer A with excess Apa I. The number of enzyme units which do not change the enzyme-specific pattern is stated in the certificate of analysis.
Absence of exonuclease activity
Approximately 5 μg [3H] labeled calf thymus DNA are incubated with 3 μl Apa I for 4 hours at +37°C in a total volume of 100 μl 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithioerythritol, pH approximately 7.5. Under these conditions, no release of radioactivity is detectable, as stated in the certificate of analysis.
Apa I recognizes the sequence GGG*CC?*C and generates fragments with 3′-cohesive termini.
특이성
Recognition sites: GGG*CC*C
GGG*CC*C
Restriction site: GGG*CC↓*C
GGG*CC↓*C
Heat inactivation: Apa I can be heat inactivated by incubation at 65 °C for 15 minutes.
GGG*CC*C
Restriction site: GGG*CC↓*C
GGG*CC↓*C
Heat inactivation: Apa I can be heat inactivated by incubation at 65 °C for 15 minutes.
DNA 프로파일
Number of cleavage sites on different DNAs
- λ: 1
- φX174: 0
- Ad2: 12
- M13mp7: 0
- pBR322: 0
- pBR328: 0
- pUC18: 0
- SV40: 1
단위 정의
One unit is the enzyme activity that completely cleaves 1 μg HindIII fragments of λDNA in one hour at +30 °C in a total volume of 25 μl (1x) SuRE/Cut Buffer A.
분석 메모
SuRE/Cut Buffer System
The buffer in bold is recommended for optimal activity
The buffer in bold is recommended for optimal activity
- A: 100%
- B: 10-25%
- H: 0-10%
- L: 50-75%
- M: 50-75%
Activity in PCR buffer: 100%
Relative activity in PCR mix (Taq DNA Polymerase buffer) is 100%. The PCR mix contained target DNA, primers, 10 mM Tris-HCl (pH 8.3, 20 °C), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 2.5 U Taq DNA polymerase. The mix was subjected to 25 amplification cycles.Activity in reaction buffer of Pwo SuperYield DNA Polymerase PCR Mix is 10%. When supplemented with GC-RICH Solution activity is increased to > 100%. Pwo SuperYield DNA Polymerase PCR Mix is not available in U.S.
Relative activity in PCR mix (Taq DNA Polymerase buffer) is 100%. The PCR mix contained target DNA, primers, 10 mM Tris-HCl (pH 8.3, 20 °C), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 2.5 U Taq DNA polymerase. The mix was subjected to 25 amplification cycles.Activity in reaction buffer of Pwo SuperYield DNA Polymerase PCR Mix is 10%. When supplemented with GC-RICH Solution activity is increased to > 100%. Pwo SuperYield DNA Polymerase PCR Mix is not available in U.S.
기타 정보
For life science research only. Not for use in diagnostic procedures.
키트 구성품 전용
제품 번호
설명
- Enzyme Solution
- SuRE/Cut Buffer A 10x concentrated
문서
The term “Restriction enzyme” originated from the studies of Enterobacteria phage λ (lambda phage) in the laboratories of Werner Arber and Matthew Meselson.
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.