추천 제품
생물학적 소스
bacterial (Brevibacterium linens)
Quality Level
형태
solution
특이 활성도
10000 U/mL
포장
pkg of 1,000 U (11558170001 [10 U/μl])
pkg of 200 U (11558161001 [10 U/μl])
제조업체/상표
Roche
파라미터
37 °C optimum reaction temp.
색상
colorless
pH
8.1 (39 °F)
solubility
water: miscible
적합성
suitable for molecular biology
응용 분야
life science and biopharma
sample preparation
외래 활성
Endonucleases, none detected (up to 20 U with MWM II-DNA)
Endonucleases, none detected (up to 20U with pBR 322-DNA)
배송 상태
dry ice
저장 온도
−20°C
일반 설명
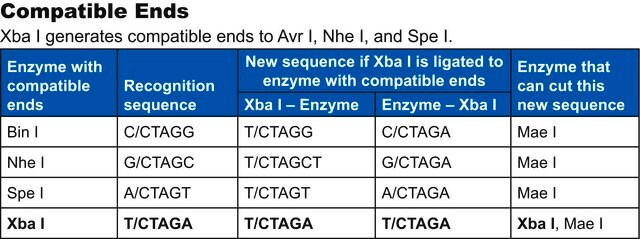
Compatible ends
Bln I ends are compatible with ends generated by Nhe I, Spe I and Xba I.
Isoschizomers
Bln I is an isoschizomer of Avr II.
Note: The complete 13 site Avr II restriction map of the E.coli genome has been reported.
Methylation sensitivity
The enzyme is not known to be affected by methylation.
특이성
CCTAGG
Restriction site: C↓CTAGG
C↓CTAGG
Heat inactivation: No inactivation of Bln I after incubation at 65 °C for 15 minutes.
품질
1 μg λDNA is incubated for 16 hours in 50 μl SuRE/Cut Buffer H with an excess of Bln I. The number of enzyme units which do not change the enzyme-specific pattern is stated in the certificate of analysis.
Absence of exonuclease activity
Approximately 5 μg [3H] labeled calf thymus DNA are incubated with 3 μl Bln I for 4 hours at +37°C in a total volume of 100 μl 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithioerythritol, pH approximately 7.5. Under these conditions, no release of radioactivity is detectable, as stated in the certificate of analysis.
Typical ligation and recutting assay
Bln I fragments obtained by complete digestion of 1 μg λ × EcoR I DNA ligated for 16 hours at +4°C with 1 U T4 DNA Ligase in 10 μl buffer that contains 66 mM Tris-HCl, 5 mM MgCl2, 5 mM Dithiothreitol, 1 mM ATP, pH 7.5 (at +20°C). The percentages of product that can be ligated and subsequently recut with Bln I and EcoR I (yielding the typical pattern of λ × EcoR I × Bln I fragments) are stated under "Lig" and "Rec" in the certificate of analysis.
DNA 프로파일
- λ: 2
- φX174: 0
- Ad2: 2
- M13mp7: 0
- M13mp18:0
- pBR322: 0
- pBR328: 0
- pUC18: 0
- SV40: 2
단위 정의
저장 및 안정성
분석 메모
Bln I has been tested in Pulsed-Field Gel Electrophoresis (on bacterial chromosomes). For cleavage of genomic DNA (E.coli C 600) embedded in agarose for PFGE analysis, we recommend using 10 U of enzyme/μg DNA and 4 hour incubation.
The buffer in bold is recommended for optimal activity
- A: 25-50%
- B: 50-75%
- H: 100%
- L: 0-10%
- M: 25-50%
기타 정보
키트 구성품 전용
- Enzyme Solution
- SuRE/Cut Buffer H 10x concentrated
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point (°F)
does not flash
Flash Point (°C)
does not flash
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
이미 열람한 고객
문서
The term “Restriction enzyme” originated from the studies of Enterobacteria phage λ (lambda phage) in the laboratories of Werner Arber and Matthew Meselson.
관련 콘텐츠
Restriction endonucleases popularly referred to as restriction enzymes, are ubiquitously present in prokaryotes. The function of restriction endonucleases is mainly protection against foreign genetic material especially against bacteriophage DNA.
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.