추천 제품
생물학적 소스
Escherichia coli ( H 560 pol A1)
Quality Level
분석
100%
형태
solution
특이 활성도
~40000 units/mg protein
포장
pkg of 100 U
제조업체/상표
Roche
기술
cDNA synthesis: suitable
색상
colorless
최적 pH
7.5-9.1
solubility
water: miscible
적합성
suitable for molecular biology
NCBI 수납 번호
응용 분야
life science and biopharma
외래 활성
RNase, none detected (up to 10 U with MS- II- RNA)
endonuclease ~10 units, none detected (using lambda-DNA)
nicking activity 10 units, none detected
배송 상태
dry ice
저장 온도
−20°C (−15°C to −25°C)
유전자 정보
Escherichia coli ... rnhA(946955)
일반 설명
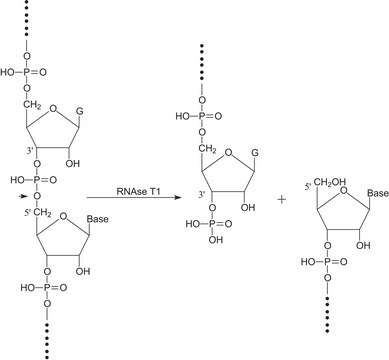
Nonspecific endoribonuclease that specifically cleaves RNA in RNA:DNA hybrids. A minimum of four continuous base pairs (RNA:DNA) is required for activity. RNase H cleaves RNA to release 5′-oligoribonucleotides.
Source: E. coli H560 pol A1
Storage Buffer: 25 mM Tris-HCl, 50 mM KCl, 1 mM dithiothreitol, 0.1 mM EDTA, 50% glycerol (v/v), pH 8.0 (+4°C)
Volume Activity: 1 x 103 U/ml assayed according to Hillenbrand & Staudenbauer.
Source: E. coli H560 pol A1
Storage Buffer: 25 mM Tris-HCl, 50 mM KCl, 1 mM dithiothreitol, 0.1 mM EDTA, 50% glycerol (v/v), pH 8.0 (+4°C)
Volume Activity: 1 x 103 U/ml assayed according to Hillenbrand & Staudenbauer.
Ribonuclease H (RNase H) is a nonspecific endoribonuclease, localized to the nucleus and cytoplasm. It is ubiquitously found and widely present among many organisms including viruses and human.
애플리케이션
Ribonuclease H (RNase H) has been used for:
- In vivo RNA-primed initiation of DNA synthesis
- Elimination of mRNA during second-strand cDNA synthesis
- Site-specific cleavage of RNA
- Detection of RNA:DNA regions in double-stranded DNA of natural origin
- Removal of poly (A) sequences of mRNA if oligo (dT) is present
- RNA extraction and quantitative reverse transcriptase polymerase chain reaction (RT-PCR)
생화학적/생리학적 작용
Ribonuclease H (RNase H) specifically cleaves RNA in RNA:DNA hybrids. A minimum of four continuous base pairs (RNA:DNA) is required for activity. RNase H cleaves RNA to release 5′-oligoribonucleotides. RNase H is associated with nucleic acid immunity. Using RNase H for degrading mRNA results in 80% depletion of mRNA and protein expression. RNase H recognizes the start codon and the 3′ and 5′ untranslated regions. This enzyme participates in DNA replication.
특징 및 장점
- Eliminate potential sources of PCR errors.
- Increase accessibility of primers during subsequent PCR.
품질
Absence of endonucleases, nicking activities, and ribonucleases.
단위 정의
RNase H is assayed according to Hillenbrand and Staudenbauer. One unit of RNase H is the amount of enzyme which produces 1 nmol acid-soluble ribonucleotides from[3H] poly(A) x poly(dT) in 20 minutes at +37 °C under the stated assay conditions.
Volume Activity: Approximately 1 U/μl
Volume Activity: Approximately 1 U/μl
제조 메모
Activator: The enzyme has its maximal activity in presence of SH-reagents
저장 및 안정성
Store at -15–-25 °C. (unopened)
기타 정보
For life science research only. Not for use in diagnostic procedures. Using RNase H after the cDNA synthesis step can increase the sensitivity of a two-step RT-PCR assay.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point (°F)
does not flash
Flash Point (°C)
does not flash
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
이미 열람한 고객
Amandine Bonnet et al.
Molecular cell, 67(4), 608-621 (2017-08-02)
Transcription is a source of genetic instability that can notably result from the formation of genotoxic DNA:RNA hybrids, or R-loops, between the nascent mRNA and its template. Here we report an unexpected function for introns in counteracting R-loop accumulation in
Antisense oligonucleotides and rna interference
Kher G, et al.
Chalcogenide Lett null
β-catenin activity in late hypertrophic chondrocytes locally orchestrates osteoblastogenesis and osteoclastogenesis.
Houben A, et al.
Development, dev-137489 (2016)
David A Ellis et al.
PLoS genetics, 17(8), e1009784-e1009784 (2021-09-01)
Aberrant repair of DNA double-strand breaks can recombine distant chromosomal breakpoints. Chromosomal rearrangements compromise genome function and are a hallmark of ageing. Rearrangements are challenging to detect in non-dividing cell populations, because they reflect individually rare, heterogeneous events. The genomic
Anirudh Chakravarthy et al.
Life science alliance, 4(12) (2021-10-02)
The continued resurgence of the COVID-19 pandemic with multiple variants underlines the need for diagnostics that are adaptable to the virus. We have developed toehold RNA-based sensors across the SARS-CoV-2 genome for direct and ultrasensitive detection of the virus and
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.