추천 제품
생물학적 소스
rabbit
Quality Level
항체 형태
affinity isolated antibody
항체 생산 유형
primary antibodies
클론
polyclonal
정제법
affinity chromatography
종 반응성
chicken, human, mouse
기술
ChIP: suitable (ChIP-seq)
dot blot: suitable
multiplexing: suitable
western blot: suitable
NCBI 수납 번호
UniProt 수납 번호
배송 상태
wet ice
타겟 번역 후 변형
acetylation (Lys9)
유전자 정보
human ... HIST1H3F(8968)
일반 설명
Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the ′beads on a string′ structure. The N-terminal tail of histone H3 protrudes from the globular nucleosome core and can undergo several different types of epigenetic modifications that influence cellular processes. These modifications include the covalent attachment of methyl or acetyl groups to lysine and arginine amino acids and the phosphorylation of serine or threonine. Acetylation of histone H3 occurs at several different lysine positions in the histone tail and is performed by a family of enzymes known as Histone Acetyl Transferases (HATs). Acetylation at lysine 9 is believed to play a role in histone deposition and chromatin assembly.
특이성
Broad species cross-reactivity is expected.
This antibody recognizes Histone H3 when acetylated at Lys9.
면역원
Epitope: Acetylated Lys9
KLH-conjugated linear peptide corresponding to human Histone H3 acetylated at Lys9.
애플리케이션
Anti-acetyl Histone H3 (Lys9) Antibody is a rabbit polyclonal antibody for detection of Histone H3 acetylated at lysine 9. This purified antibody, also known as Anti-H4K9ac has dot blot (DB) proven specificity and has been validated in WB, DB, ChIP, Mplex.
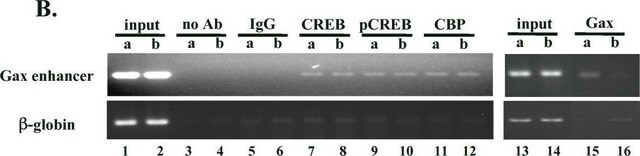
Chromatin Immunoprecipitation Analysis (ChIP):
A representative lot of this antibody immunoprecipitated chromatin from Histone H3 acetylated on Lys9.
ChIP-Sequencing:
Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 μg of a representative lot of anti-acetyl-Histone H3 (Lys9) antibody (ABE18), 20μL Protein A/G beads, and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of thirteen million reads from FastQ files were mapped using Bowtie(http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files.
Dot Blot Analysis:
A representative lot pf this antibody detected Histone H3 in modified and non modified peptides of acetylated and non-acetylated Histone H3 Lys9.
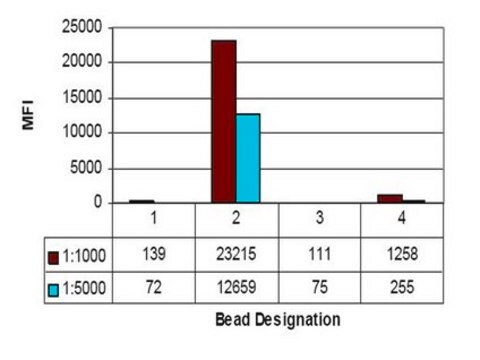
Luminex Analysis:
A representative lot of this antibody specifically recognized Histone H3 acetylated on Lys9 by Luminex assay.
A representative lot of this antibody immunoprecipitated chromatin from Histone H3 acetylated on Lys9.
ChIP-Sequencing:
Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 μg of a representative lot of anti-acetyl-Histone H3 (Lys9) antibody (ABE18), 20μL Protein A/G beads, and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of thirteen million reads from FastQ files were mapped using Bowtie(http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files.
Dot Blot Analysis:
A representative lot pf this antibody detected Histone H3 in modified and non modified peptides of acetylated and non-acetylated Histone H3 Lys9.
Luminex Analysis:
A representative lot of this antibody specifically recognized Histone H3 acetylated on Lys9 by Luminex assay.
Research Category
Epigenetics & Nuclear Function
Epigenetics & Nuclear Function
Research Sub Category
Histones
Histones
품질
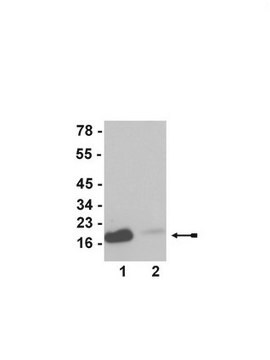
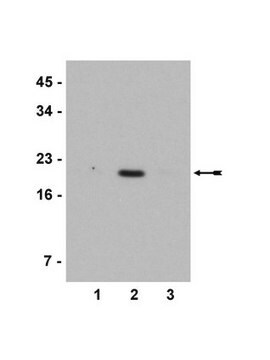
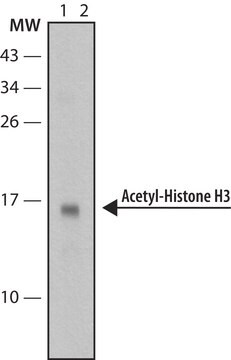
Evaluated by Western Blot in sodium butyrate untreated and treated HeLa acid extract.
Western Blot Analysis: 0.05 µg/mL of this antibody detected Histone H3 on 10 µg of sodium butyrate untreated and treated HeLa acid extract.
Western Blot Analysis: 0.05 µg/mL of this antibody detected Histone H3 on 10 µg of sodium butyrate untreated and treated HeLa acid extract.
표적 설명
~17 kDa observed
물리적 형태
Affinity purified
Purified rabbit polyclonal in buffer containing 0.1 M Tris-Glycine (pH 7.4), 150 mM NaCl with 0.05% sodium azide.
저장 및 안정성
Stable for 1 year at 2-8°C from date of receipt.
분석 메모
Control
Sodium butyrate untreated and treated HeLa acid extract
Sodium butyrate untreated and treated HeLa acid extract
기타 정보
Concentration: Please refer to the Certificate of Analysis for the lot-specific concentration.
면책조항
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
적합한 제품을 찾을 수 없으신가요?
당사의 제품 선택기 도구.을(를) 시도해 보세요.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point (°F)
Not applicable
Flash Point (°C)
Not applicable
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
Isaac K Sundar et al.
American journal of physiology. Lung cellular and molecular physiology, 311(6), L1245-L1258 (2016-10-30)
Chromatin-modifying enzymes mediate DNA methylation and histone modifications on recruitment to specific target gene loci in response to various stimuli. The key enzymes that regulate chromatin accessibility for maintenance of modifications in DNA and histones, and for modulation of gene
Simone Roeh et al.
Nucleus (Austin, Tex.), 8(4), 370-380 (2017-04-28)
Different types of sequencing biases have been described and subsequently improved for a variety of sequencing systems, mostly focusing on the widely used Illumina systems. Similar studies are missing for the SOLiD 5500xl system, a sequencer which produced many data
Fei Liu et al.
PLoS pathogens, 9(9), e1003613-e1003613 (2013-09-27)
Covalently closed circular DNA (cccDNA) of hepadnaviruses exists as an episomal minichromosome in the nucleus of infected hepatocyte and serves as the transcriptional template for viral mRNA synthesis. Elimination of cccDNA is the prerequisite for either a therapeutic cure or
Bernard Chi Hang Lee et al.
Cell reports, 14(8), 1819-1828 (2016-02-26)
Centromeres, the specialized chromosomal regions for recruiting kinetochores and directing chromosome segregation, are epigenetically marked by a centromeric histone H3 variant, CENP-A. To maintain centromere identity through cell cycles, CENP-A diluted during DNA replication is replenished. The licensing factor M18BP1(KNL-2)
Linyuan Peng et al.
Nucleic acids research, 48(9), 4992-5005 (2020-04-03)
SIRT6 deacetylase activity improves stress resistance via gene silencing and genome maintenance. Here, we reveal a deacetylase-independent function of SIRT6, which promotes anti-apoptotic gene expression via the transcription factor GATA4. SIRT6 recruits TIP60 acetyltransferase to acetylate GATA4 at K328/330, thus
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.