추천 제품
관련 카테고리
일반 설명
All ChIPAb+ antibodies are individually validated for chromatin precipitation, every lot, every time. Each ChIPAb+ antibody set includes control primers (tested every lot by qPCR) to biologically validate your IP results in a locus-specific context. The qPCR protocol and primer sequences are provided, allowing researchers to validate ChIP protocols when using our antibody in their chromatin context. Each set also includes a negative control antibody to ensure specificity of the ChIP reaction.
The ChIPAb+ Dimethyl-Histone H3 (Lys4) set includes the Anti-dimethyl-Histone H3 (Lys4) antibody, a negative control antibody (purified Mouse IgG), and qPCR primers which amplify a 213 bp region within the coding region of the human GAPDH gene. The dimethyl-histone H3 (Lys4) and negative control antibodies are supplied in a scalable "per ChIP" reaction size and can be used to functionally validate the precipitation of dimethyl-histone H3 (Lys4)-associated chromatin.
The ChIPAb+ Dimethyl-Histone H3 (Lys4) set includes the Anti-dimethyl-Histone H3 (Lys4) antibody, a negative control antibody (purified Mouse IgG), and qPCR primers which amplify a 213 bp region within the coding region of the human GAPDH gene. The dimethyl-histone H3 (Lys4) and negative control antibodies are supplied in a scalable "per ChIP" reaction size and can be used to functionally validate the precipitation of dimethyl-histone H3 (Lys4)-associated chromatin.
특이성
Recognizes histone H3, Mr 17 kDa, dimethylated at lysine 4.
The peptide sequence is identical in a wide range of animal and plant species, so broad cross-reactivity is expected.
면역원
Epitope: a.a. 1-12
The dimethyl-histone H3 (Lys4) purified antibody is made against a synthetic peptide (dimethylated at Lys4) corresponding to amino acids 1-12 of histone H3.
애플리케이션
Chromatin Immunoprecipitation:
Sonicated chromatin prepared from HeLa cells (2 X 106 cell equivalents per IP) was subjected to chromatin immunoprecipitation using 2 µg of either normal mouse IgG, or Anti-dimethyl-Histone H3 (Lys4) antibody and the Magna ChIP G Kit (Cat. #17-611). Successful immunoprecipitation of dimethyl-histone H3 (Lys4)-associated DNA fragments was verified by qPCR using β-globin ChIP Primers versus GAPDH Coding primers (Please see figures). Data is presented as percent input of each IP sample relative to input chromatin for each antibody with the indicated primers.
Please refer to the EZ-Magna G ChIP (Cat. #17-408) or EZ-ChIP (Cat. #17-371) protocol for experimental details.
ChIP-seq Analysis:
Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 µg Anti-dimethyl-Histone H3 (Lys4) antibody (cat# 17-677), 20 µL Protein A/G beads, and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of sixteen million reads from FastQ files were mapped using Bowtie (http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files. The highest 25% of peaks identified in the 04-790 and 17-677 datasets showed 92 and 90% overlap with peaks identified in the ENCODE H3K4me2 BROAD Histone track for HeLa S3.
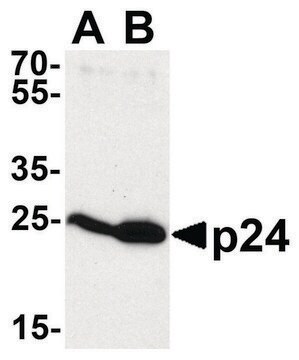
Western Blot Analysis:
Recombinant Histone H3 (Lane 1) and HeLa acid extract (Lane 2) were resolved by electrophoresis, transferred to PVDF membrane and probed with Anti-dimethyl Histone H3 (Lys4) (2 μg/mL).
Proteins were visualized using a goat anti-mouse secondary antibody conjugated to HRP (Cat. #AP124P) and a chemiluminescence detection system.
Dot Blot Analysis:
Absurance Histone H3 Antibody Specificity Array (Cat. No. 16-667) and Absurance Histone H2A, H2B, H4 Antibody Specificity Array (Cat. No. 16-665), which contain histone peptides with various modifications were probed with Anti-dimethyl H3 (Lys4) at 2.0ug/mL (1:500) dilution. Proteins were visualized using a Donkey anti-mouse IgG conjugated to HRP and a chemiluminescence detection system.
Beadlyte Histone Peptide Specificity Assay:
0.15 μg/ml of purified anti-dimethyl-Histone H3 (Lys4),
clone CMA303 was incubated with a cocktail of microspheres
conjugated to histone H3 peptides with the following modifications:
1. unmodified H3 (K4)
2. monomethyl H3 (K4)
3. dimethyl H3 (K4)
4. trimethyl H3 (K4)
5. trimethy H3 (K27)
Unbound antibody was then removed by filtration.
Peptide antibody complexes were incubated with a
PE-conjugated anti-mouse secondary antibody.
Fluorescence was read on a Luminex 100
instrument. Median Fluorescence intensity (MFI) is plotted.
Sonicated chromatin prepared from HeLa cells (2 X 106 cell equivalents per IP) was subjected to chromatin immunoprecipitation using 2 µg of either normal mouse IgG, or Anti-dimethyl-Histone H3 (Lys4) antibody and the Magna ChIP G Kit (Cat. #17-611). Successful immunoprecipitation of dimethyl-histone H3 (Lys4)-associated DNA fragments was verified by qPCR using β-globin ChIP Primers versus GAPDH Coding primers (Please see figures). Data is presented as percent input of each IP sample relative to input chromatin for each antibody with the indicated primers.
Please refer to the EZ-Magna G ChIP (Cat. #17-408) or EZ-ChIP (Cat. #17-371) protocol for experimental details.
ChIP-seq Analysis:
Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 µg Anti-dimethyl-Histone H3 (Lys4) antibody (cat# 17-677), 20 µL Protein A/G beads, and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of sixteen million reads from FastQ files were mapped using Bowtie (http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files. The highest 25% of peaks identified in the 04-790 and 17-677 datasets showed 92 and 90% overlap with peaks identified in the ENCODE H3K4me2 BROAD Histone track for HeLa S3.
Western Blot Analysis:
Recombinant Histone H3 (Lane 1) and HeLa acid extract (Lane 2) were resolved by electrophoresis, transferred to PVDF membrane and probed with Anti-dimethyl Histone H3 (Lys4) (2 μg/mL).
Proteins were visualized using a goat anti-mouse secondary antibody conjugated to HRP (Cat. #AP124P) and a chemiluminescence detection system.
Dot Blot Analysis:
Absurance Histone H3 Antibody Specificity Array (Cat. No. 16-667) and Absurance Histone H2A, H2B, H4 Antibody Specificity Array (Cat. No. 16-665), which contain histone peptides with various modifications were probed with Anti-dimethyl H3 (Lys4) at 2.0ug/mL (1:500) dilution. Proteins were visualized using a Donkey anti-mouse IgG conjugated to HRP and a chemiluminescence detection system.
Beadlyte Histone Peptide Specificity Assay:
0.15 μg/ml of purified anti-dimethyl-Histone H3 (Lys4),
clone CMA303 was incubated with a cocktail of microspheres
conjugated to histone H3 peptides with the following modifications:
1. unmodified H3 (K4)
2. monomethyl H3 (K4)
3. dimethyl H3 (K4)
4. trimethyl H3 (K4)
5. trimethy H3 (K27)
Unbound antibody was then removed by filtration.
Peptide antibody complexes were incubated with a
PE-conjugated anti-mouse secondary antibody.
Fluorescence was read on a Luminex 100
instrument. Median Fluorescence intensity (MFI) is plotted.
Research Category
Epigenetics & Nuclear Function
Epigenetics & Nuclear Function
Research Sub Category
Chromatin Biology
Chromatin Biology
This ChIPAb+ Dimethyl-Histone H3 (Lys4) -ChIP Validated Antibody & Primer Set conveniently includes the antibody & the specific control PCR primers.
포장
25 assays per kit, ~2μg per chromatin immunoprecipitation
품질
Chromatin Immunoprecipitation:
Sonicated chromatin prepared from HeLa cells (1 X 106 cell equivalents per IP) were subjected to chromatin immunoprecipitation using 2 µg of either a normal mouse IgG, or Anti-dimethyl-Histone H3 (Lys4) antibody and the Magna ChIP G Kit (Cat. #17-611). Successful immunoprecipitation of dimethyl-histone H3 (Lys4) associated DNA fragments was verified by qPCR using ChIP Primers GAPDH Coding (Please see figures).
Please refer to the EZ-Magna G ChIP (Cat. #17-409) or EZ-ChIP (Cat. #17-371) protocol for experimental details.
Sonicated chromatin prepared from HeLa cells (1 X 106 cell equivalents per IP) were subjected to chromatin immunoprecipitation using 2 µg of either a normal mouse IgG, or Anti-dimethyl-Histone H3 (Lys4) antibody and the Magna ChIP G Kit (Cat. #17-611). Successful immunoprecipitation of dimethyl-histone H3 (Lys4) associated DNA fragments was verified by qPCR using ChIP Primers GAPDH Coding (Please see figures).
Please refer to the EZ-Magna G ChIP (Cat. #17-409) or EZ-ChIP (Cat. #17-371) protocol for experimental details.
표적 설명
Dimethyl-histone H3 at ~17 kDa
물리적 형태
Anti-dimethyl-Histone H3 (Lys4) (mouse monoclonal IgG1, clone CMA303). One vial containing 50 μg of protein G purified antibody in 50 μL PBS containing 0.05% sodium azide. Store at -20°C.
Normal Mouse IgG. Two vials containing 25 μg purified Mouse IgG in 25 μL storage buffer containing 0.1% sodium azide. Store at -20°C.
ChIP Primers GAPDH Coding. One vial containing 75 μL of 5 μM of each primer specific for the coding region human GAPDH. Store at -20°C.
FOR: GGC TCC CAC CTT TCT CAT CC
REV: GGC CAT CCA CAG TCT TCT GG
Normal Mouse IgG. Two vials containing 25 μg purified Mouse IgG in 25 μL storage buffer containing 0.1% sodium azide. Store at -20°C.
ChIP Primers GAPDH Coding. One vial containing 75 μL of 5 μM of each primer specific for the coding region human GAPDH. Store at -20°C.
FOR: GGC TCC CAC CTT TCT CAT CC
REV: GGC CAT CCA CAG TCT TCT GG
Format: Purified
저장 및 안정성
Stable for 1 year at -20°C from date of receipt.
Aliquot upon initial thaw, avoid freeze thaw cycles.
Aliquot upon initial thaw, avoid freeze thaw cycles.
분석 메모
Control
Includes negative control mouse IgG antibody and control primers specific for human β-globin promoter.
Includes negative control mouse IgG antibody and control primers specific for human β-globin promoter.
법적 정보
UPSTATE is a registered trademark of Merck KGaA, Darmstadt, Germany
면책조항
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Storage Class Code
10 - Combustible liquids
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
Arpana Verma et al.
Journal of cellular biochemistry, 116(5), 743-753 (2014-12-17)
In this study genome-wide di-methylated H3K4 (H3K4me2) and tri-methylated H3K27 (H3K27me3) modification profiles were analyzed in spermatozoa of buffalo bulls having wide fertility differences. The custom designed 4 × 180 K buffalo (Bubalus bubalis) ChIP-on-chip array was fabricated by employing array-based sequential hybridization
Xuncheng Liu et al.
Plant physiology, 158(1), 119-129 (2011-10-14)
The molecular mechanism of how the histone deacetylase HDA6 participates in maintaining transposable element (TE) silencing in Arabidopsis (Arabidopsis thaliana) is not yet defined. In this study, we show that a subset of TEs was transcriptionally reactivated and that TE
Xuhong Yu et al.
Current biology : CB, 31(23), 5377-5384 (2021-10-20)
Transcription initiation has long been considered a primary regulatory step in gene expression. Recent work, however, shows that downstream events, such as transcription elongation, can also play important roles.1-3 A well-characterized example from animals is promoter-proximal pausing, where transcriptionally engaged
Anand Balasubramani et al.
PLoS genetics, 10(1), e1003969-e1003969 (2014-01-15)
Differentiation-dependent regulation of the Ifng cytokine gene locus in T helper (Th) cells has emerged as an excellent model for functional study of distal elements that control lineage-specific gene expression. We previously identified a cis-regulatory element located 22 kb upstream
Xiaojie Li et al.
Proceedings of the National Academy of Sciences of the United States of America, 109(28), 11425-11430 (2012-06-27)
REPRESSOR OF SILENCING 1 (ROS1) is a DNA demethylation enzyme that was previously identified during a genetic screen for the silencing of both RD29A-LUC and 35S-NPTII transgenes on a T-DNA construct. Here we performed a genetic screen to identify additional
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.