Degrader Building Blocks with Inhibitor of Apoptosis Protein (IAP) In Silico-Derived Ligands
Introduction to Targeted Protein Degradation with IAP In Silico-Derived Ligands
Targeted protein degradation (TPD) is an emerging strategy that uses small molecules to hijack endogenous proteolysis systems to degrade disease-relevant proteins and thus reduce their abundance in cells. The small molecules are called protein degraders, which are heterobifunctional and contain two ligands connected by a crosslinker – a warhead directed at the target protein and a ligand for a cellular E3 ubiquitin ligase protein (Figure 1A).1

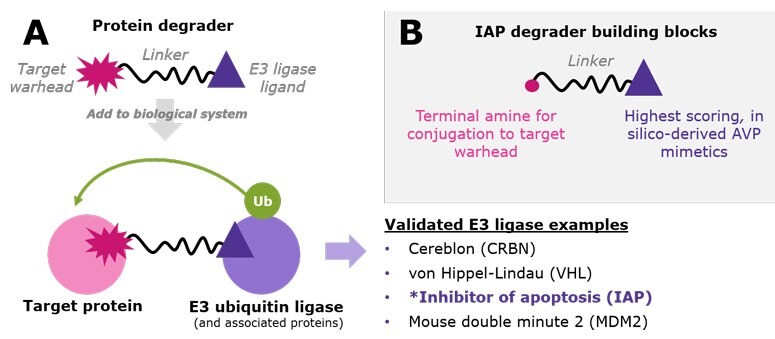
Figure 1.A.) Protein degraders are bifunctional small molecules that recruit a target protein to an E3 ubiquitin ligase. This proximity facilitates polyubiquitination of the target protein, resulting in its proteasomal degradation. While the list is growing, examples of validated E3 ligases used in TPD research include CRBN, VHL, IAP, and MDM2. B.) IAP degrader building blocks incorporate three novel virtual screening leads already conjugated to varied linkers with amine-terminal chemistry.
The E3 ligases are the gates to the proteasomal degradation system, and characterization and small-molecule targeting of the >600 in the human proteome is an active area of research.2–3 For now, a handful of E3 ligases are most widely used. Proteolysis-targeting chimeras (PROTAC® degraders) have been developed for cereblon (CRBN) and von Hippel–Lindau (VHL) E3 ligases (Figure 1A),1–3 and our original degrader building blocks streamline the synthesis of CRBN and VHL-mediated degrader libraries. Other degraders such as specific and non-genetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs) recruit IAP ubiquitin ligases.4–5 To offer novel chemistry in this space, we have partnered with ComInnex to develop IAP degrader building blocks containing three novel, in silico-derived IAP lead compounds (Figure 1B). Each is conjugated at the N- or C-terminus with both long and short aliphatic and PEG-based linkers (Figure 2). Together, these reagents accelerate synthesis of IAP-mediated degraders at early stages of TPD research and assessment. Learn more about the computational discovery of the IAP ligands below.
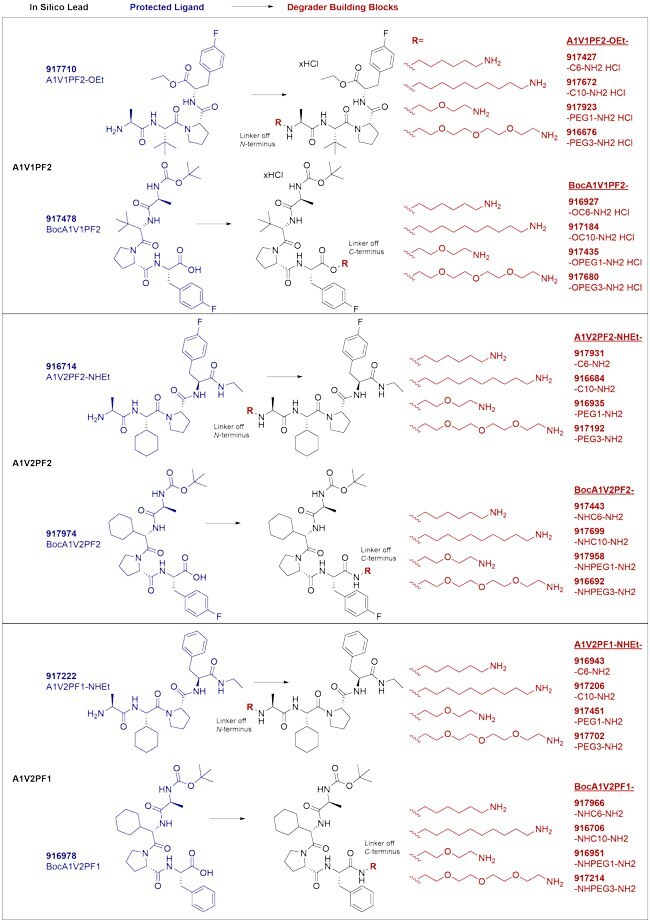
Figure 2.IAP-targeted ligands and degrader building blocks based on in silico screening of AVP mimetics. In silico AVP lead ligands are shown at the left; each is conjugated at the N- or C-terminus with both a long and short aliphatic and PEG-based linker to streamline synthesis of degraders and thus assessment of IAP-mediated degradation utilizing this chemistry.
Advantages of IAP In Silico-Derived Ligands
- Investigate the validated IAP E3 ligase in your protein degrader development
- Access novel, in silico-derived, IAP-targeted AVP leads via virtual screening
- Streamline synthesis of potential IAP-mediated protein degraders using AVP leads already conjugated to linkers
- Obtain upfront glimpse of SAR with varied exit vectors and linkers
- Simplify parallel synthesis with consistent amide terminal chemistry among the IAP degrader building blocks as well as those for CRBN and VHL
AVPI-Based Ligand Discovery
The IAP family contains various proteins with high similarity6 (XIAP: X-linked IAP, cIAP1/2: cellular IAP1/2). XIAP inhibits initiator (Caspase-9) and effector caspases (Caspase-3 and -7) through the interaction of its BIR domains (BIR3 and BIR2) with the N-terminal tetrapeptide (Ala-Val-Pro-Ile, AVPI) of the SMAC protein (Second Mitochondria-derived Activator of Caspases). Similarly, AVPI is able to bind to the BIR3 domain of cIAPs, and this interaction induces auto-degradation of the protein through ubiquitination (Ub). However, if AVPI mimetics are linked to warheads specific to proteins of interest, the conjugate could induce ubiquitination and degradation of target proteins.
Based on the structure of AVPI (Ala-Val-Pro-Ile), systematic research was initiated exploring the chemical space7 and led to the discovery of potent apoptosis-inducing IAP antagonists (AVPF, MV1). While the natural AVPI tetrapeptide had moderate binding activities towards XIAP-BIR3 (Kd = AVPI: 580 nM); introducing bulky hydrophobic groups8 readily increased the activity (AVPF [AVPI Ile -> Phe] Kd = 290 nM). Furthermore, the related N-Me-AVPF-NH2 analogue showed increased activity towards cIAP1 BIR3 domains (IC50 XIAP = 108.2 nM, cIAP1 = 48.2 nM; cIAP2 = 209 nM).9 The continuation of this trend led to the identification of the highly potent diphenylmethyl-containing pan-IAP antagonist, MV1 (Kd values: XIAP BIR3 = 16 nM, cIAP1 BIR3: 5.8 nM)).10 (Figure 3)
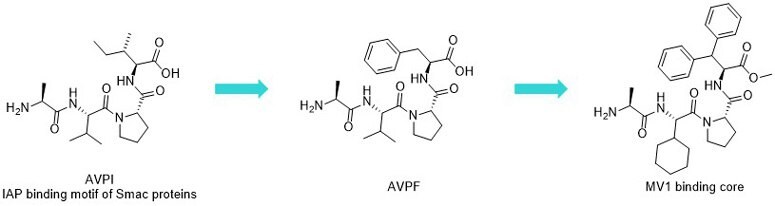
Figure 3.AVPI (Ala-Val-Pro-Ile) and IAP antagonists AVPF and MV1.
These compounds also act as E3 ligase-triggering small molecules, thus having potential for development of protein degraders as demonstrated with SNIPERs and PROTACs.5 Conjugated to various target-binding small molecules, these chimeras are capable of degrading various proteins of interest (POIs; cellular retinoic acid binding protein (CRABP-II), estrogen receptor-α (ERα), androgen receptor (AR), BCR-ABL protein kinase, transforming acidic coiled-coil-3 (TACC3) and Halo-tag proteins).5, 11
AVPI mimetics provide potential building blocks for the generation and development of efficient PROTAC libraries.
Virtual Screening (VS) of the AVPI-Mimetics Library
Based on the structure of AVPF and MV1, an AVPI-mimetics library was generated (Figure 4A) for in silico screening, and a resulting AVP-based compound library of potentially active ligands were selected according to docking scores.
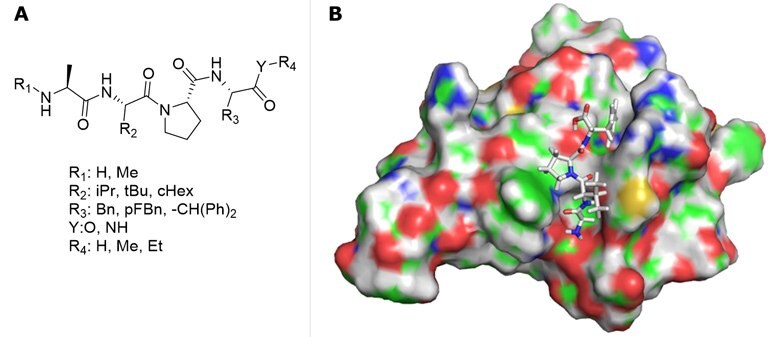
Figure 4.A) AVPI-mimetics library diversification for virtual screening. B) Representative docking of AVPF.
Schrödinger "Small Molecule Drug Discovery Suite" software package was used for virtual screening.12 To incorporate flexibility of the receptor, the Induced Fit Docking (IFD) module was applied, followed by MM-GBSA binding free energy calculations. High-resolution cIAP1 BIR3 structures were collected from PDB (4kmn, 3uw4, 4hy4, 4hy5, 4mti, 4mu7), and cross docking calculations were performed by docking all ligands into all 3D structures followed by MM-GBSA ΔGbind calculations. The 4lge structure13 provided the highest average binding free energy; therefore, this structure was used for further calculations. Representative docking structure of AVPF is illustrated in Figure 4B. IFD docking results are summarized in Table 1, confirming previous activity trends and indicating that the new derivatives (entries 4-6) have improved binding potency.
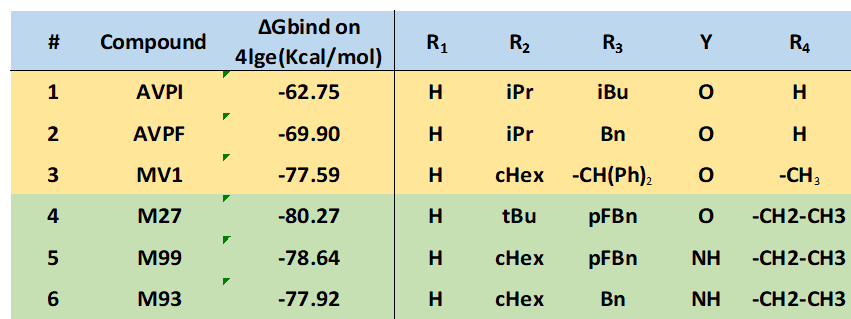
Table 1.cIAP BIR3 docking scores (ΔGbind values).
The selected molecules have two functional groups: an amino group in alanine (N-terminal) and a carboxyl function at the other end of the tetrapeptide (C-terminal) that makes the compounds attractive for further derivatization. To extend to development of novel degrader compounds, modelling calculations confirmed linker conjugation at both ends (Figure 5). For synthetic purposes, the compounds were orthogonally protected by Boc groups.
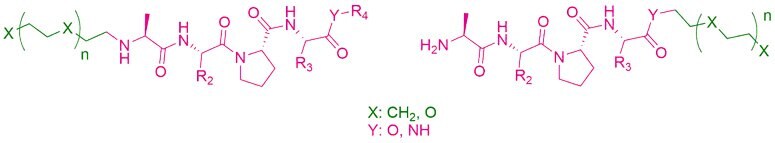
Figure 5.AVP-based lead compounds are turned into degrader building blocks through linker conjugation at the N- or C-terminus.
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
Special thanks to ComInnex for jointly preparing this technology spotlight!
References
続きを確認するには、ログインするか、新規登録が必要です。
アカウントをお持ちではありませんか?