SAE0045
HRV-3C Protease.
N-Terminal His tagged recombinant protein, aqueous solution, 0.8-1.2 mg/mL
Sinonimo/i:
Human Rhinovirus 3C Protease, Levlfqgp site protease, PreScission Protease
About This Item
Prodotti consigliati
Origine biologica
human (human Rhinovirus Type 14)
Livello qualitativo
Saggio
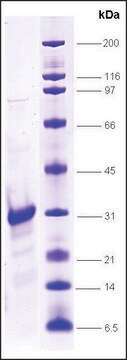
≥90% (SDS-PAGE)
Forma fisica
aqueous solution
Attività specifica
≥5000 U/mg
PM
21 kDa
Concentrazione
0.8-1.2 mg/mL
tecniche
protein purification: suitable
Compatibilità
suitable for protein modification
applicazioni
life science and biopharma
Condizioni di spedizione
dry ice
Temperatura di conservazione
−20°C
Descrizione generale
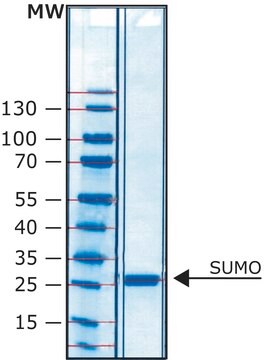
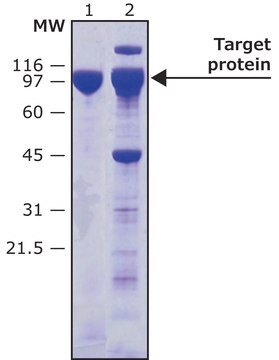
Proteolytic cleavage occurs between the Gln and Gly residues.
HRV-3C protease is a useful tool to cleave recombinant proteins that are expressed as a fusion protein with this sequence, between the carrier domain and the protein of interest.
This recombinant version contains a six-histidine tag and can be easily removed by IMAC chromatography.
Applicazioni
Definizione di unità
Stato fisico
Codice della classe di stoccaggio
10 - Combustible liquids
Classe di pericolosità dell'acqua (WGK)
WGK 2
Certificati d'analisi (COA)
Cerca il Certificati d'analisi (COA) digitando il numero di lotto/batch corrispondente. I numeri di lotto o di batch sono stampati sull'etichetta dei prodotti dopo la parola ‘Lotto’ o ‘Batch’.
Possiedi già questo prodotto?
I documenti relativi ai prodotti acquistati recentemente sono disponibili nell’Archivio dei documenti.
I clienti hanno visto anche
Il team dei nostri ricercatori vanta grande esperienza in tutte le aree della ricerca quali Life Science, scienza dei materiali, sintesi chimica, cromatografia, discipline analitiche, ecc..
Contatta l'Assistenza Tecnica.