SAB4200625
Anti-Methyl-Histone H3 (Me-Lys9)(H3K9me1) antibody, Mouse monoclonal
clone 7E7-H12, purified from hybridoma cell culture
Synonyme(s) :
9430068D06RIK, H3.3A, H3.3B, H3F3, H3F3A, H3F3A/H3F3B, H3F3B, HISTONE 3B, LOC100045490, RP11−396C23.1, wu:fb58e10, zgc:56193, zgc:86731
About This Item
Produits recommandés
Source biologique
mouse
Niveau de qualité
Conjugué
unconjugated
Forme d'anticorps
purified immunoglobulin
Type de produit anticorps
primary antibodies
Clone
7E7-H12, monoclonal
Forme
buffered aqueous solution
Poids mol.
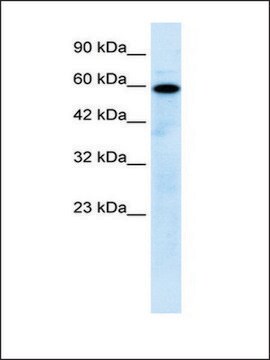
antigen ~17 kDa
Espèces réactives
human
Concentration
~1 mg/mL
Technique(s)
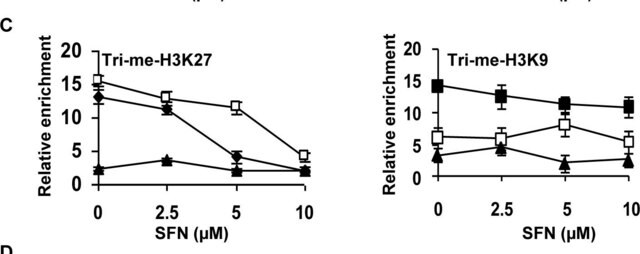
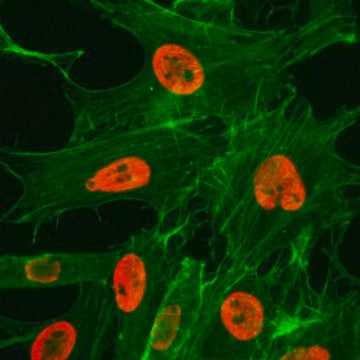
immunoblotting: 2.5-5 μg/mL using human HeLa cells.
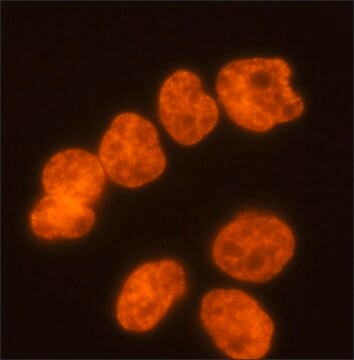
immunocytochemistry: 2.5-5 μg/mL using human HeLa cells.
Isotype
IgG1
Conditions d'expédition
dry ice
Température de stockage
−20°C
Modification post-traductionnelle de la cible
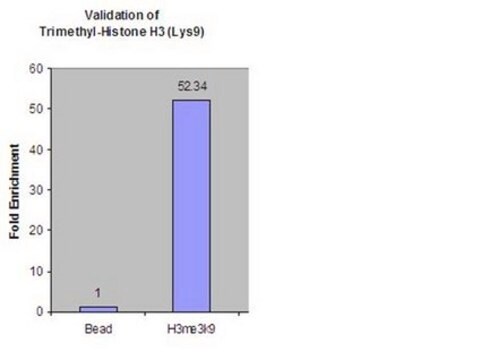
monomethylation (Lys9)
Informations sur le gène
human ... H3C1(8350)
Description générale
Immunogène
Application
Actions biochimiques/physiologiques
Forme physique
Clause de non-responsabilité
Vous ne trouvez pas le bon produit ?
Essayez notre Outil de sélection de produits.
Code de la classe de stockage
10 - Combustible liquids
Classe de danger pour l'eau (WGK)
WGK 1
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Faites votre choix parmi les versions les plus récentes :
Certificats d'analyse (COA)
Vous ne trouvez pas la bonne version ?
Si vous avez besoin d'une version particulière, vous pouvez rechercher un certificat spécifique par le numéro de lot.
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique