908401
1-Methyl-7-nitroisatoic anhydride
Synonyme(s) :
1-Methyl-7-nitro-2H-3,1-benzoxazine-2,4(1H)-dione, 1-methyl-7-nitro-2H-3,1-Benzoxazine-2,4(1H)-dione, 1M7, RNA SHAPE probe, Reagent for RNA SHAPE-MaP
About This Item
Produits recommandés
Forme
powder
Pf
204.5 °C
Température de stockage
2-8°C
InChI
1S/C9H6N2O5/c1-10-7-4-5(11(14)15)2-3-6(7)8(12)16-9(10)13/h2-4H,1H3
Clé InChI
MULNCJWAVSDEKJ-UHFFFAOYSA-N
Application
Autres remarques
SnapShot: RNA Structure Probing Technologies
Detection of RNA-Protein Interactions in Living Cells with SHAPE
Standardization of RNA Chemical Mapping Experiments
In-cell RNA structure probing with SHAPE-MaP
A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry
Produit(s) apparenté(s)
Code de la classe de stockage
11 - Combustible Solids
Classe de danger pour l'eau (WGK)
WGK 3
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique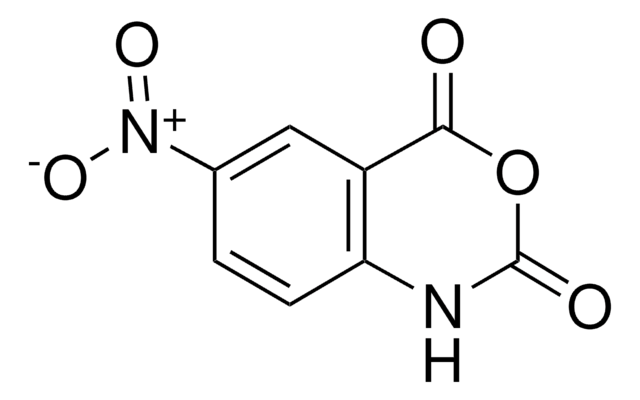
![1,4-Diazabicyclo[2.2.2]octane ReagentPlus®, ≥99%](/deepweb/assets/sigmaaldrich/product/structures/366/129/a6ff4175-974d-4fac-9038-b35e508ef252/640/a6ff4175-974d-4fac-9038-b35e508ef252.png)
![1,8-Diazabicyclo[5.4.0]undéc-7-ène 98%](/deepweb/assets/sigmaaldrich/product/structures/120/564/5b373e23-1624-489c-8efb-692de0f96ffb/640/5b373e23-1624-489c-8efb-692de0f96ffb.png)