N1290
Polynucleotide phosphorylase human
Synonym(s):
Polyribonucleotide nucleotidyltransferase
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
recombinant
expressed in E. coli
form
solution
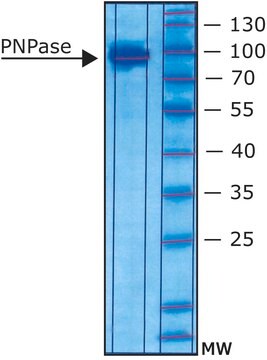
enzyme activity
≥20 units/mg protein
mol wt
~90 kDa
concentration
400-600 μg/mL protein
shipped in
dry ice
storage temp.
−70°C
Biochem/physiol Actions
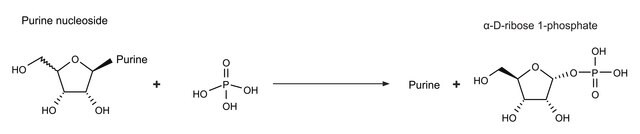
Polynucleotide phosphorylase (PNPase) is a bifunctional enzyme with a phosphorolytic 3′ to 5′ exoribonuclease activity and a 3′-terminal oligonucleotide polymerase activity. It is also involved in mRNA processing and degradation in bacteria, plants, and humans.
Physical form
supplied as a solution in 20 mM HEPES buffer, pH 7.9, with 0.1 mM EDTA, 2 mM DTT, 12.5 mM MgCl2, ~130 mM KCl, and 20% (w/v) glycerol.
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Lukasz S Borowski et al.
Biochimica et biophysica acta, 1797(6-7), 1066-1070 (2010-02-02)
Protein complexes responsible for RNA degradation play important role in three key aspects of RNA metabolism: they control stability of physiologically functional transcripts, remove the unnecessary RNA processing intermediates and destroy aberrantly formed RNAs. In mitochondria the post-transcriptional events seem
Victoria Portnoy et al.
RNA (New York, N.Y.), 14(2), 297-309 (2007-12-18)
PNPase is a major exoribonuclease that plays an important role in the degradation, processing, and polyadenylation of RNA in prokaryotes and organelles. This phosphorolytic processive enzyme uses inorganic phosphate and nucleotide diphosphate for degradation and polymerization activities, respectively. Its structure
Hsiao-Wen Chen et al.
Molecular and cellular biology, 26(22), 8475-8487 (2006-09-13)
We recently identified polynucleotide phosphorylase (PNPase) as a potential binding partner for the TCL1 oncoprotein. Mammalian PNPase exhibits exoribonuclease and poly(A) polymerase activities, and PNPase overexpression inhibits cell growth, induces apoptosis, and stimulates proinflammatory cytokine production. A physiologic connection for
Takashi Nagaike et al.
The Journal of biological chemistry, 280(20), 19721-19727 (2005-03-17)
Mammalian mitochondrial (mt) mRNAs have short poly(A) tails at their 3' termini that are post-transcriptionally synthesized by mt poly(A) polymerase (PAP). The polyadenylation of mt mRNAs is known to be a key process needed to create UAA stop codons that
Magdalena Leszczyniecka et al.
Proceedings of the National Academy of Sciences of the United States of America, 99(26), 16636-16641 (2002-12-11)
Terminal differentiation and cellular senescence display common properties including irreversible growth arrest. To define the molecular and ultimately the biochemical basis of the complex physiological changes associated with terminal differentiation and senescence, an overlapping-pathway screen was used to identify genes
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service