N9914
Polynucleotide phosphorylase from Synechocystis sp.
recombinant, expressed in E. coli
Synonym(s):
PNPase, Polyribonucleotide Nucleotidyltransferase
Sign Into View Organizational & Contract Pricing
All Photos(2)
About This Item
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
biological source
bacterial (Synechocystis sp.)
Quality Level
recombinant
expressed in E. coli
description
Histidine tagged
Assay
90% (SDS-PAGE)
form
solution
specific activity
≥500 units/mg protein
mol wt
85 kDa
technique(s)
cell based assay: suitable
suitability
suitable for molecular biology
application(s)
cell analysis
shipped in
dry ice
storage temp.
−70°C
Looking for similar products? Visit Product Comparison Guide
General description
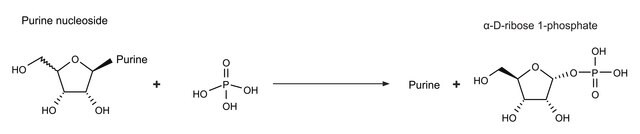
Polynuclotide phosphorlyase in spinach chloroplasts acts as a exonuclease and a poly(A) polymerase.
Application
Polynucleotide phosphorylase has been used in a study to discover that a major function of PNPase is the synthesis of CDP. It has also been used in a study to investigate the enzyme responsible for RNA 3′-tail synthesis in S. coelicolor.
Biochem/physiol Actions
Polynucleotide phosphorylase (PNPase) is a bifunctional enzyme with a phosphorolytic 3′ to 5′ exoribonuclease activity and a 3′-terminal oligonucleotide polymerase activity.
Polynucleotide phosphorylase localizes to the intermembrane space of mitochondria and has a critical function in regulating mitochondrial homeostasis in human cells.
Unit Definition
One unit will polymerize 1.0 μmole of ADP, releasing 1.0 μmole of inorganic phosphate in 15 minutes, at pH 9.1 at 37 °C.
Supplied as a solution in 20 mM Hepes buffer pH 7.9, 0.1 mM EDTA, 2 mM DTT, 12.5 mM MgCl2, 60 mM KCl, 20% (w/v) Glycerol
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Patricia Bralley et al.
Microbiology (Reading, England), 152(Pt 3), 627-636 (2006-03-04)
As in other bacteria, 3'-tails are added post-transcriptionally to Streptomyces coelicolor RNA. These tails are heteropolymeric, and although there are several candidates, the enzyme responsible for their synthesis has not been definitively identified. This paper reports on three candidates for
Ruth Rott et al.
The Journal of biological chemistry, 278(18), 15771-15777 (2003-02-26)
The mechanism of RNA degradation in Escherichia coli involves endonucleolytic cleavage, polyadenylation of the cleavage product by poly(A) polymerase, and exonucleolytic degradation by the exoribonucleases, polynucleotide phosphorylase (PNPase) and RNase II. The poly(A) tails are homogenous, containing only adenosines in
A Danchin
DNA research : an international journal for rapid publication of reports on genes and genomes, 4(1), 9-18 (1997-02-28)
Genome comparison permits identification of chromosome regions conserved during evolution. Bacillus subtilis and Escherichia coli are so distant that there exists very few conserved landmarks in their genome organisation. Analysis of the conserved cmk rpsA cluster pinpointed the importance of
G G Liou et al.
Proceedings of the National Academy of Sciences of the United States of America, 98(1), 63-68 (2001-01-03)
RNase E isolated from Escherichia coli is contained in a multicomponent "degradosome" complex with other proteins implicated in RNA decay. Earlier work has shown that the C-terminal region of RNase E is a scaffold for the binding of degradosome components
Peter Lengyel
Annual review of microbiology, 66, 27-38 (2012-09-22)
2011 marked the fiftieth anniversary of breaking the genetic code in 1961. Marshall Nirenberg, the National Institutes of Health (NIH) scientist who was awarded the Nobel Prize in Physiology or Medicine in 1968 for his role in deciphering the code
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service