MSP09
Membrane Scaffold Protein 1E3D1 D73C
recombinant, expressed in E. coli, Cysteine substituted at position 73
Iniciar sesiónpara Ver la Fijación de precios por contrato y de la organización
About This Item
Código UNSPSC:
12352200
NACRES:
NA.26
Productos recomendados
recombinante
expressed in E. coli
Ensayo
≥90% (SDS-GE)
Formulario
buffered aqueous solution
Condiciones de envío
ambient
temp. de almacenamiento
−20°C
Descripción general
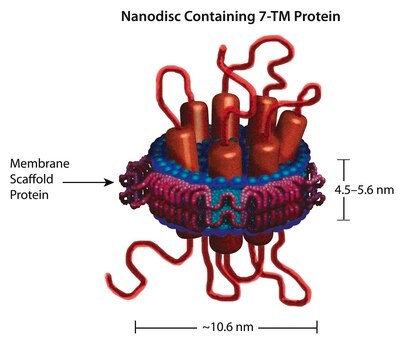
Nanodisc technology is an approach to render membrane proteins soluble in aqueous solutions in a native-like bilayer environment, where the membrane proteins remain stable and active. The Nanodisc concept is derived from high density lipoprotein (HDL) particles and their primary protein component, apolipoprotein. The Nanodisc is a non-covalent structure of phospholipid bilayer and membrane scaffold protein (MSP), a genetically engineered protein, which mimics the function of Apolipoprotein A-1 (ApoA-1).
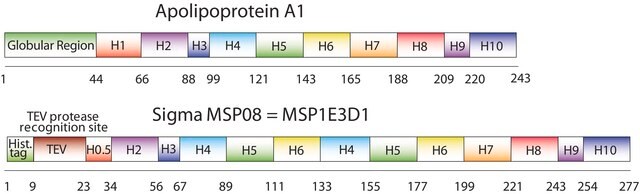
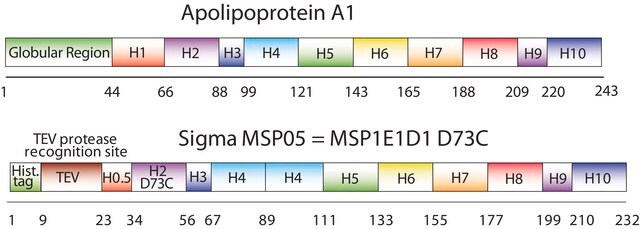
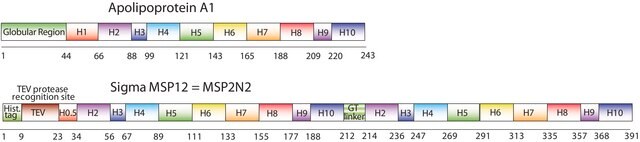
The first MSP, MSP1, was engineered with its sequence based on the sequence of A-1, but without the globular N-terminal domain of native A-1. The MSP1E3D1 D73C variant of MSP1 differs from MSP1 in the following facets:
The first MSP, MSP1, was engineered with its sequence based on the sequence of A-1, but without the globular N-terminal domain of native A-1. The MSP1E3D1 D73C variant of MSP1 differs from MSP1 in the following facets:
- It deletes the first 11 amino acids in the Helix 1 portion (referred to as “H0.5” in the accompanying figure) of the original MSP1 sequence3 (which is known separately as MSP1D1).
- It repeats the Helix 4 (H4), Helix 5 (H5) and Helix 6 (H6) sequences of the original MSP1 sequence between the parent Helix 6 (H6) and Helix 7 (H7) segments of MSP1D1.
- It substitutes a cysteine (C) residue for an aspartic acid (D) residue in the Helix 2 (H2) portion of the protein, at position 73 of the original native A-1 sequence.
- The initial histidine-tag is one amino acid shorter.
Aplicación
For guidelines on the use of this and other MSP′s to prepare Nanodiscs, please visit our Protocols for Membrane Scaffold Proteins and Nanodisc Formation page.
Información legal
Nanodisc technology, and many of its uses, are covered by the following patents held by the University of Illinois.
- 7,691,414 Membrane scaffold proteins
- 7,662,410 Membrane scaffold proteins and embedded membrane proteins
- 7,622,437 Tissue factor compositions and methods
- 7,592,008 Membrane scaffold proteins
- 7,575,763 Membrane scaffold proteins and tethered membrane proteins
- 7,083,958 Membrane scaffold proteins
- 7,048,949 Membrane scaffold proteins
Código de clase de almacenamiento
12 - Non Combustible Liquids
Clase de riesgo para el agua (WGK)
WGK 2
Punto de inflamabilidad (°F)
Not applicable
Punto de inflamabilidad (°C)
Not applicable
Elija entre una de las versiones más recientes:
Certificados de análisis (COA)
Lot/Batch Number
¿No ve la versión correcta?
Si necesita una versión concreta, puede buscar un certificado específico por el número de lote.
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Rory N Pruitt et al.
The New phytologist, 215(2), 725-736 (2017-05-31)
The biotrophic pathogen Xanthomonas oryzae pv. oryzae (Xoo) produces a sulfated peptide named RaxX, which shares similarity to peptides in the PSY (plant peptide containing sulfated tyrosine) family. We hypothesize that RaxX mimics the growth-stimulating activity of PSY peptides. Root length
Yunting Pu et al.
Frontiers in plant science, 8, 1204-1204 (2017-07-27)
Autophagy is a critical process for recycling of cytoplasmic materials during environmental stress, senescence and cellular remodeling. It is upregulated under a wide range of abiotic stress conditions and is important for stress tolerance. Autophagy is repressed by the protein
Protocolos
Protocols for Membrane Scaffold Proteins and Nanodisc Formation
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico