SHC005
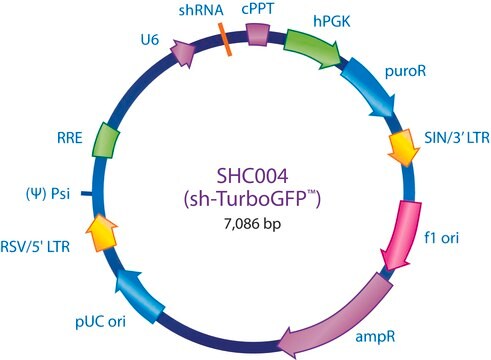
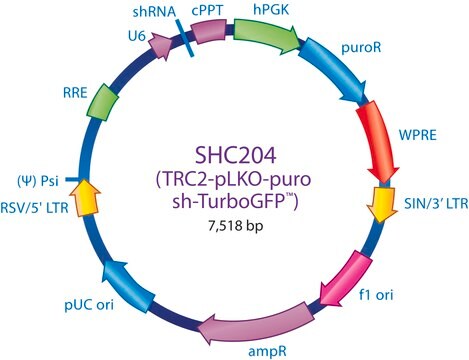
MISSION® eGFP shRNA Control Vector
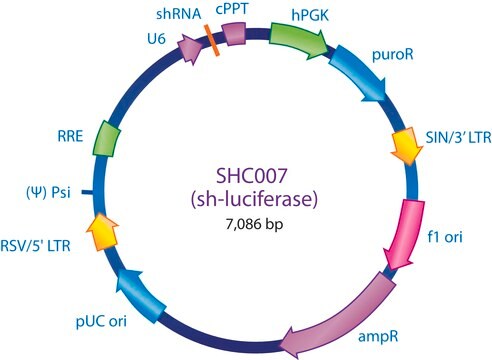
shRNA sequence targeting eGFP
Synonyme(s) :
MISSION® Control Vectors
About This Item
Produits recommandés
Niveau de qualité
Gamme de produits
MISSION®
Concentration
500 ng/μL in TE buffer; DNA (10μg of plasmid DNA)
Conditions d'expédition
dry ice
Température de stockage
−20°C
Vous recherchez des produits similaires ? Visite Guide de comparaison des produits
Catégories apparentées
Description générale
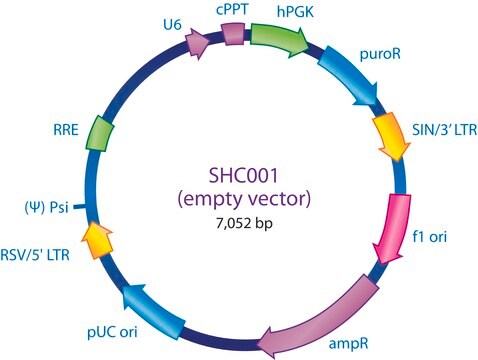
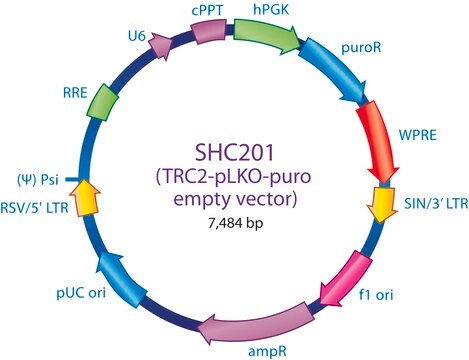
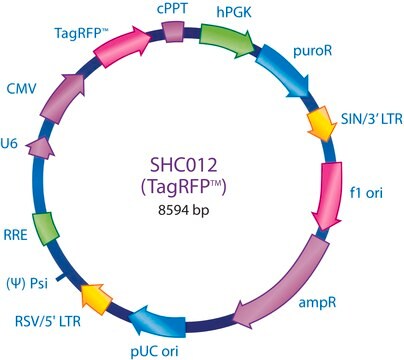
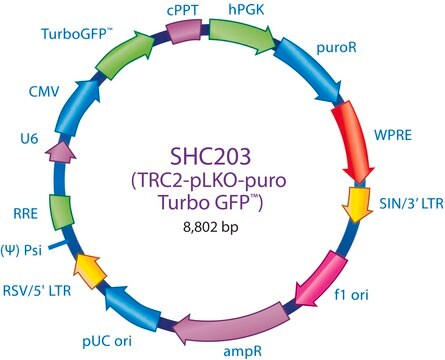
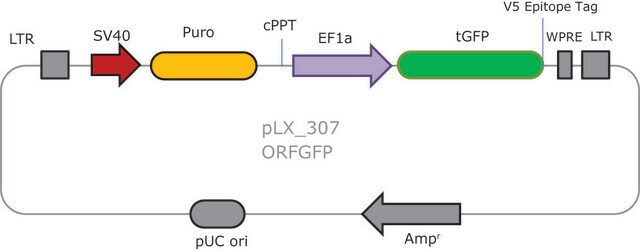
Ampicillin and puromycin antibiotic resistance genes provide selection in bacterial or mammalian cells respectively. In addition, self-inactivating replication incompetent viral particles can be produced in packaging cells (HEK293T) by co-transfection with compatible packaging plasmids, MISSION® Lentiviral Packaging Mix (Prod. No. SHP001). The eGFP shRNA Control Vector is provided as 10 μg of plasmid DNA in Tris-EDTA (TE) buffer at a concentration of 500 ng/μl.
Application
Informations légales
En option
Code de la classe de stockage
12 - Non Combustible Liquids
Classe de danger pour l'eau (WGK)
WGK 1
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Les clients ont également consulté
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique