Wichtige Dokumente
P5568
Proteinase K aus Tritirachium album
≥500 units/mL, buffered aqueous glycerol solution
Synonym(e):
Endopeptidase K
About This Item
Empfohlene Produkte
Biologische Quelle
microbial (T.album
T. ALBUM)
Qualitätsniveau
Form
buffered aqueous glycerol solution
Mol-Gew.
28.93 kDa
Konzentration
≥10 mg/mL
≥500 units/mL
Methode(n)
DNA extraction: suitable
Lagertemp.
2-8°C
Suchen Sie nach ähnlichen Produkten? Aufrufen Leitfaden zum Produktvergleich
Allgemeine Beschreibung
Anwendung
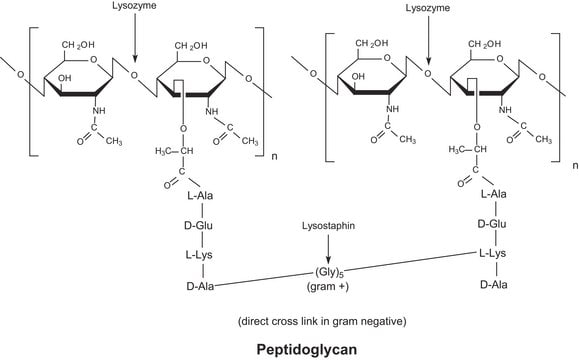
Entfernt Endotoxine, die an kationische Proteine wie Lysozym und Ribonuklease A binden.
Berichten zufolge nützlich für die Isolierung von Leber-, Hefe- und Mungbohnen-Mitochondrien
Bestimmung der Enzymlokalisierung auf Membranen
Behandlung von Paraffinschnitten zur Freilegung von Antigenbindungsstellen zur Antikörpermarkierung.
Verdauung von Proteinen aus Hirngewebeproben für Prionen in der TSE-Forschung.
- to break down cardiac muscle during histopathology studies
- during the digestion of HEK-293 cells
Biochem./physiol. Wirkung
Einheitendefinition
Physikalische Form
Angaben zur Herstellung
Signalwort
Danger
H-Sätze
P-Sätze
Gefahreneinstufungen
Resp. Sens. 1
Lagerklassenschlüssel
10 - Combustible liquids
WGK
WGK 1
Flammpunkt (°F)
Not applicable
Flammpunkt (°C)
Not applicable
Persönliche Schutzausrüstung
Eyeshields, Faceshields, Gloves, type ABEK (EN14387) respirator filter
Analysenzertifikate (COA)
Suchen Sie nach Analysenzertifikate (COA), indem Sie die Lot-/Chargennummer des Produkts eingeben. Lot- und Chargennummern sind auf dem Produktetikett hinter den Wörtern ‘Lot’ oder ‘Batch’ (Lot oder Charge) zu finden.
Besitzen Sie dieses Produkt bereits?
In der Dokumentenbibliothek finden Sie die Dokumentation zu den Produkten, die Sie kürzlich erworben haben.
Kunden haben sich ebenfalls angesehen
Artikel
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Protokolle
Proteinase K (EC 3.4.21.64) activity can be measured spectrophotometrically using hemoglobin as the substrate. Proteinase K hydrolyzes hemoglobin denatured with urea, and liberates Folin-postive amino acids and peptides. One unit will hydrolyze hemoglobin to produce color equivalent to 1.0 μmol of tyrosine per minute at pH 7.5 at 37 °C (color by Folin & Ciocalteu's Phenol Reagent).
Unser Team von Wissenschaftlern verfügt über Erfahrung in allen Forschungsbereichen einschließlich Life Science, Materialwissenschaften, chemischer Synthese, Chromatographie, Analytik und vielen mehr..
Setzen Sie sich mit dem technischen Dienst in Verbindung.